In this article we will discuss about the concept of wobble hypothesis.
Crick (1966) proposed the ‘wobble hypothesis’ to explain the degeneracy of the genetic code. Except for tryptophan and methionine, more than one codons direct the synthesis of one amino acid. There are 61 codons that synthesise amino acids, therefore, there must be 61 tRNAs each having different anticodons. But the total number of tRNAs is less than 61.
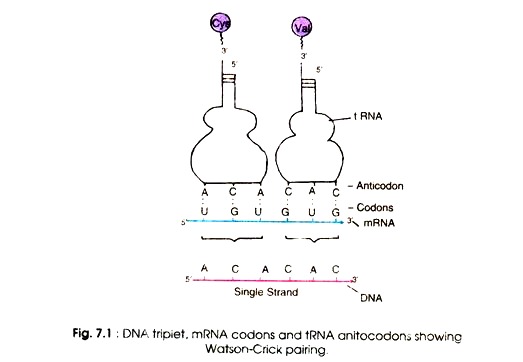
This may be explained that the anticodons of some tRNA read more than one codon. In addition, identity of the third codon seems to be unimportant. For example CGU, CGC, CGA and CGG all code for arginine. It appears that CG specifies arginine and the third letter is not important. Conventionally, the codons are written from 5′ end to 3′ end.
Therefore, the first and second bases specify amino acids in some cases. According to the Wobble hypothesis, only the first and second bases of the triple codon on 5′ → ‘3 mRNA pair with the bases of the anticodon of tRNA i.e A with U, or G with C.
The pairing of the third base varies according to the base at this position, for example G may pair with U. The conventional pairing (A = U, G = C) is known as Watson-Crick pairing (Fig. 7.1) and the second abnormal pairing is called wobble pairing.
This was observed from the discovery that the anticodon of yeast alanine-tRNA contains the nucleoside inosine (a deamination product of adenosine) in the first position (5′ → 3′) that paired with the third base of the codon (5′ → 3′). Inosine was also found at the first position in other tRNAs e.g. isoleucine and serine.
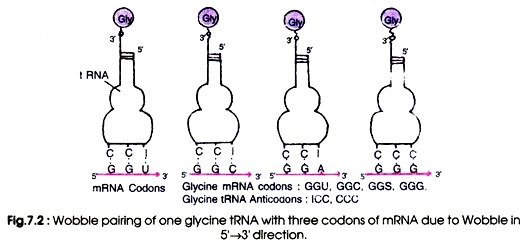
The purine, inosine, is a wobble nucleotide and is similar to guanine which normally pairs with A, U and C. For example a glycine-tRNA with anticodon 5′-ICC-3′ will pair with glycine codons GGU, GGC, GGA and GGG (Fig 7.2). Similarly, a seryl-tRNA with anticodon 5′-IGA-3′ pairs with serine codons UCC, UCU and UCA (5-3′). The U at the wobble position will be able to pair with an adenine or a guanine.
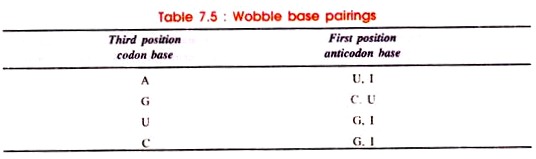
According to Wobble hypothesis, allowed base pairings are given in Table 7.5:
Due to the Wobble base pairing one tRNA becomes able to recognise more than one codons for an individual amino acid. By direct sequence of several tRNA molecules, the wobble hypothesis is confirmed which explains the pattern of redundancy in genetic code in some anticodons (e.g. the anticodons containing U, I and G in the first position in 5’→ 3′ direction)
The seryl-tRNA anticodon (UCG) 5′-GCU-3′ base pairs with two serine codons, 5′-AGC-3′ and 5′-AGU-3′. Generally, Watson-Crick pairing occurs between AGC and GCU. However, in AGU and GCU pairing, hydrogen bonds are formed between G and U. Such abnormal pairing called ‘Wobble pairing’ is given in Table 7.5.
Three types of wobble pairings have been proposed:
(i) U in the wobble position of the tRNA anticodon pairs with A or G of codon,
(ii) G pairs with U or C, and
(iii) 1 pairs with A, U or C.