Transposons are of two types, composite transposon and complex transposon.
1. The Composite Transposons:
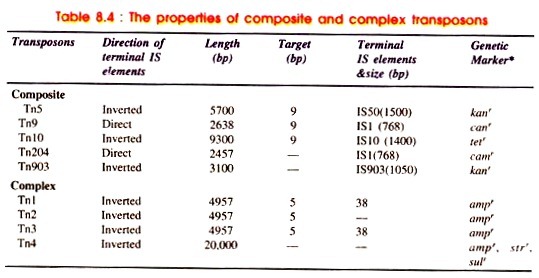
The composite transposons are those which consist of a central region carrying antibiotic resistant genes flanked at both the ends by identical copies of an IS element Therefore, composite transposons carry drug resistance or other markers in addition to transposition (Fig. 8.32A). It is a class of larger transposons. Three frequently studied composite transposons are Tn5, Tn9, and Tn10.
Tn5 element shows kanamycin resistance (kanr) and consists of 5,400 base pair segment with 1,450 bp inverted repeats at both ends of the segment. It can be transposed from phage λ to the chromosome of E .coli and from a locus of K coli chromosome to another locus. If inserted into genes it causes mutations.
Tn9 transposon consists of an R factor derived gene for chloramphemcol resistance (camr). The enzyme that confers drug resistance consists of 2,638 sequences in the middle of Tn flanked by 768 bp long IS1 element on either side. Tn9 IS1 is present in direct order with small inverted repeat at ends.
The camr segment is translocated from an R-factor to F-episome as well as phage λ through the phage P1. T9 differs from the other transposons in its instability and a high frequency loss of its antibiotic resistance can be encountered.
Tn10 consists of tetracycline resitance genes (tetr genes). It is 9,300 bp long consisting of inverted repeats, on either side of 1,400 base pairs. The IS elements is IS10. Tn10 can be translocated from R222 (a drug resitance plasmid) to phage P22.
Structure of composite transposons:
In the composite transposons, the IS elements can be in an inverted or direct repeat configuration (Fig. 8.31B). The two ends of IS elements are themselves inverted repeats. The relative orientation (direct or inverted) of the flanking IS elements of a composite transposon does not alter its terminal sequences. Thus a composite transposon with arms of direct repeats has the structure: arm L- central region – arm R.
The structure becomes: arm L-central region – arm R, if the arms are inverted repeats. The arrows show the orientation of arms according to the orientation of genetic map of the transposon from left (L) to right (R) (Fig. 8.31B, i-ii). The features of composite transposons are given in Table 8.4.
Moreover, there are some cases where modules of a composite transposon are identical, for example Tn9 (has direct repeat of IS1) or Tn903 (inverted repeats of IS 903 present), whereas in certain cases the modules are closely related. Hence, the modules in Tn10 or Tn5 can be distinguished. However, when the modules are identical both can sponsor the movement of transposon as found in Tn9 of Tn903.
When the modules differ, they may also differ in functional ability. Therefore, transposition depends entirely on one of the modules for example Tn10 or Tn5. Thus an IS module when functional, transposes either itself or the whole transposon.
The ability of a single module to transpose the entire composite transposon explains the lack of selective pressure for both the modules to remain active. The IS elements code for transposase activity that is responsible both for creating a target site and for recognizing the ends of transposon. Only the ends are needed for a transposon to serve as a substrate for transposition.
2. The Complex Transposons (TnA transposon family):
The TnA family of transposons includes Tn1, Tn2, and Tn3. TnA consists of quite large elements (about 500 bp). These contain independent units carrying genes for transposition as well as for drug resistance. These are not composite relaying on IS-type transposition modules. The TnA family includes several related transposons of which Tn3 and Tn10 are the best studied ones.
TnA family is also known with the name Tn3 family because Tn3 transposon was first discovered in bacterial plasmids in 1974 by Hedges and Jacob. Tn3 is an example of a complex transposon. It takes a modular structure as found in Tn10 and is not based on IS elements. Also, it does not has evolutionary links to IS elements. The basic structure of Tn3 is described for TnA family.
One of the unique features of TnA family is that it limits multiple insertions of the same transposon into a plasmid. Most surprising finding is that the transposition frequency to other plasmids in the same cell is unaffected. Heffron (1979) have given the analysis of DNA sequence of transposon Tn3.
Structure of Transposons of TnA family:
Transposons of TnA family consist of inverted terminal repeats (of 38 bp long but none are flanked by IS-like elements), an internal res site and three known genes e.g. tnpA, tnpR and ampr.
The gene tnpA encodes for transposase and tnpR encodes for resolvase. The ampr gene (5 bp long) is generated at the target site as direct repeat and codes for β-lactamase that confers resistance to ampicillin. The res site consists of three subunits: I, II and III (Fig. 8.33)