The tRNA, mRNA and rRNA are involved in the process of transcription. However, an enzyme transcriptase i.e. DNA-dependent RNA polymerase is required for the synthesis of RNA by using ribonucleotide triphosphates i.e ATP, GTP, CTP and UTP. Structure and function of RNA polymerase are described herewith.
Contents
1. RNA Polymerase:
On a DNA template elongation of RNA chain at each step is catalysed by RNA polymerase. RNA polymerase is found both in prokaryotes and eukaryotes but the structure and function in these two groups of organisms differ.
In prokaryotes only a single enzyme, RNA polymerase governs the synthesis of all cellular RNAs, whereas in eukaryotes, several types of RNA polymerases are involved for the synthesis of a cellular RNA. For example, mRNA, tRNA and rRNA in E. coli are synthesized by the same RNA polymerase.
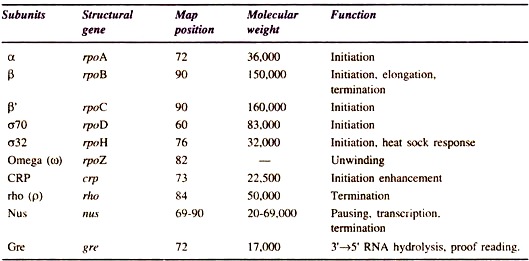
The holoenzyme is a complete RNA polymerase consisting of core, an enzyme and a sigma (σ) factor, hence it is a complex enzyme. The core enzyme of E. coli consists of five polypeptide chains, two α subunits, one (β), one β’ subunits and one omega (ω) subunit (Fig. 10.2). There are seven polypeptide chains (e.g. β αα δω1, ω2) in core enzyme of E. coli. The holoenzyme binds to DNA at specific sites called promoters and transcribes specific length of RNA.
Thus o factor plays a significant role in promoter recognition by RNA polymerase. It can easily be isolated from the holoenzyme. The β subunit consists of a catalytic site for RNA synthesis and binding sites for substrates and products as well. The β subunit plays a role in binding RNA polymerase to the DNA template.
The two a subunits assemble the two larger subunits into core enzyme (α2ββ’ω). The function of small subunit (ω) is not known in detail; however, it is supposed that it takes part in unwinding. DNA polymerase subunits, structural genes and their functions are summarized in Table 10.1. The E. coli RNA polymerase synthesizes RNA at the rate of 40 nucleotides per minutes at 37°C.
(i) Types of RNA Polymerase:
There are three different eukaryotic RNA polymerases that are transcribed by three different sets of genes (Fig. 10.3).
They are distinguished by their sensitivity to a fungal toxin, α-amanitin:
(a) RNA polymerase I (RNA Pol I):
It is located in the nucleolus and synthesizes precursors of most rRNAs. It is sensitive to α-amanitin.
(b) RNA polymerase II (RNA Pol II):
It is located in the nucleoplasm and synthesizes mRNA precursors and some small nuclear RNAs. It is very sensitive to α-amanitin.
(c) RNA polymerase III (RNA Pol III):
It is located in the nucleoplasm. It synthesizes the precursors of tRNA, 5S rRNA and other small nuclear and cytoplasmic RNAs. It is moderately sensitive to α-amanitin.
In addition, an antibiotic rifampicin commonly inhibits transcription by interfering with P subunit of prokaryotic RNA polymerase. Ishihawa (1992) has presented a current view of RNA polymerase structure and some binding sites for each subunit (Fig. 10.4).
Fig. 10.4 : Functional map of RNA polymerase subunits.
(ii) The Site of Transcription:
Transcription begins at the promoter site i.e. cistron localised on DNA molecule. Out of two only one strand of dsDNA is transcribed. It has also been demonstrated that in ØX174 for any cistron only one strand of DNA is transcribed into RNA. In addition, in phage λ and phage T4 both strands are transcribed, where one strand serves as template for some genes.
The rest of genes are transcribed from the other strand. The DNA strand that acts as template is known as antisense strand. The nucleotide sequences of anti- sense strand and mRNA transcripts are complementary. The other strand is called sense strand. The bases of sense strand and that of mRNA are exactly the same.
In some viruses e.g. SP8, ØX174, etc. out of two only one strand is transcribed. In SV40 both the strands are transcribed for different genes. In contrast in T even phages, phage λ, E. coli and eukaryotes different regions of both the DNA strand act as template for RNA synthesis.
Table 10.1 : RNA polymerase subunits and other transcription factors.
(iii) Process of Transcription:
The process of transcription is accomplished in the following three main steps: chain initiation, chain elongation and chain termination. The complete process of transcription is outlined in Fig. 10.5.
2. Chain Initiation:
During the process of transcription the components required to initiate the RNA chain are the template activated precursors, divalent metal ions (Mg++ or Mn++), RNA polymerase and template.
(i) Promoter Recognition:
The enzyme RNA polymerase plays a key role in recognition and binding of initiation site. Before the formation of complex enzyme, sigma factor interacts with core enzyme at p subunit site. This is required to check transcription of both the strands by core enzyme. The holoenzyme transcribes only one of the two DNA strands. The sigma factor of holoenzyme recognizes the promoter region of the DNA (Fig.10.6).
A specific base sequence (20-200 bases long) is called a promoter. Examination of a large number of promoter sequences of different genes from different bacteria has shown that they have many common features. The promoter sequences are centred at -10 and -35 base pairs from the transcription start point and are implicated in normal promoter function.
E. coli has the following two distinct hexamer components of promoter:
The -10 sequence is called Pribnow’s box, whereas -35 region is called the recognition site. The Pribnow’s box is found as a part of all prokaryotic promoters. The -35 region is located about 35 base pairs upstream (away from the direction of transcription site). The promoter is upstream from the structural gene.
The RNA polymerase interacts with groups in the major groove and recognises the proper sequence upstream (-35 region) from the Pribnow’s box, thereafter forms stable complex by moving laterally to the -10 region. Thus, the sigma factor recognises both the above regions.
(ii) Binding of RNA Polymerase to Promoter:
The strength of binding of RNA polymerase to different promoters varies. There are some promoters that have binding sites for proteins rather than RNA polymerase.
Therefore, for these promoters the site must be occupied by that protein for RNA polymerase to bind correctly. One of the most common binding sites for this type is one that binds a complex of cyclic AMP receptor protein (CRP). The concentration of AMP governs the activity of these promoters.
(iii) Unwinding of DNA Double Helix:
The super helical nature of the chromosome may play a role in promoter function. Generally, a negatively super coiled chromosome is a better transcriptional template than relaxed chromosome. The torsional stress imposed by super coiling makes the certain area of DNA easier to separate by RNA polymerase.
Hence, super coiling affects the expression of some genes more than the others. Binding of omega (ω) factor of RNA polymerase results in unwinding of a DNA helix. Consequently, a short segment of DNA opens. The open complex then allows tight binding of the RNA polymerase with subsequent initiation of RNA synthesis.
(iv) Synthesis of First Base of RNA Chain:
The first base of RNA synthesised is always in the form of purine i.e. either triphosphate guanine (pppG) or adenine (pppA). Most of the chains in E. coli is started with pppG, whereas in phage T7 and ØX174 it is initiated with pppA. In contrast to DNA replication, initiation of mRNA synthesis does not require primers. Initiation ends after the formation of first inter nucleotide bond (5’pppPupN) (Fig.10.5).
3. Chain Elongation:
Chain elongation occurs by the core enzyme that moves along the DNA template. After beginning the elongation, transcription goes on at a rate between 30 and 60 nucleotides per second at 37°C.
In general the elongation reaction includes the steps:
(i) Nucleotide triphosphate binding,
(ii) Bond formation between the nucleotide and the 3′-OH of nascent RNA chain,
(iii) Pyrophosphate release, and
(iv) Translocation of polymerase along the DNA template.
The activated ribonucleotide triphosphates i.e. ATP, UTP, OTP and CTP are added according to nucleotides of one strand of DNA template. The incoming nucleotide forms hydrogen bonds with a DNA base. Reaction occurs between the nucleotides at 3′ end of the RNA molecule and P in the triphosphate group leading to removal of phosphates (PPi). The PPi is soon hydrolysed to inorganic phosphate (Pi) (Fig. 10.7A).
(i) Release of Sigma (σ) Factor:
When 8-9 nucleotide long mRNA transcript is formed, the σ factor is released abruptly from the complex. Consequently this causes a decrease in the affinity of σ from the RNA polymerase-DNA-nascent RNA complex. The release of σ can combine with any of core enzyme and thus it is reused for the initiation of a new chain (Fig.10.5).
(ii) Direction of Transcription:
The ribonucleotide triphosphates attach to the first nucleotide of the template and chain growth takes place in 5′ → 3′ direction. The region of template that has been transcribed regains its double helical form behind the bubble and the next region of DNA which is to be transcribed unwinds.
The RNA transcript does not elongate uniformly along the template. This is due to the presence of pausing sites at certain regions in the template. It has been found that generally the pausing sequence contains GC rich regions about 16-20 bp, upstream of 3′-OH end of the paused transcript. Also, the region of dyad symmetry present 16-20 bp upstream of 3′-end causes pausing. However, the mechanism of pausing is not clear.
Moreover, it is believed that after release of sigma factor the NusA and NusG proteins become associated with core enzyme and modulate the rate of elongation. At some sites these proteins enhance pausing.
It is interesting to note that the newly formed RNA transcript consists of a triphosphate group at 5′ and free -OH group at 3′ end (Fig. 10.7 B). Moreover, the RNA transcript is larger than the structural gene. Most messages contain a promoter i.e. proximal region of mRNA which is called the leader sequence i.e. the un-translated region before the beginning of translation.
(iii) Proof Reading of mRNA:
The RNA polymerase possesses proof reading activity which is analogous to the 3′ → 5′ exonuclease activity of DNA polymerase. The two proteins, GreA and GreB enable the RNA polymerase (which is transcriptionally arrested) to back up and cleave 2-3 nucleotides from 3′ end of nascent message. This results in release of RNA polymerase which in turn moves through the arrest point at the 3′ end of the gene.
(iv) Protein-Protein Interaction with RNA Polymerase:
There are several regulatory factors that directly contact the RNA polymerase at the promoter region of template. Such regulatory factors (proteins) can be divided into two classes, I and II.
Class I proteins act as activator of transcription. These bind upstream from the promoter (e.g. CRP, AraC, Fnr and OmpR). Class II transcriptional factors overlap the promoter region; a few of these regulators make contact with C-terminal end of the α subunit (Fig. 10.4).
4. Chain Termination:
The process of termination of RNA chain ends with the events:
(i) Cessation of elongation,
(ii) Release of transcript from the tertiary complex, and
(iii) Dissociation of polymerase from the DNA template.
There are two types of mechanisms that bring about termination: rho-independent and rho-dependent terminations.
(i) Rho-independent Termination:
The rho-independent signal for termination is recognised by DNA itself. It consists of GC rich region with dyad symmetry. RNA polymerase reads and extends polyA sequence on DNA template (Fig.10.8A). It synthesizes an RNA transcript that becomes folded to form a stem and loop structure of about 20 bases upstream from the 3′-OH terminus with a terminal stretch of 4-8 poly U residues (Fig. 10.8B).
This RNA stem-loop structure causes RNA polymerase to pause and disrupt the RNA-DNA hybrid at 5′ end. Poly U residues are present at 3′ end of RNA-DNA hybrid molecule. The U-A hybrid base pairs are relatively unstable; therefore, it causes the 3′ end of the hybrid to break and release the mRNA chain.
(ii) Rho-dependent Termination:
Richardson (1983) has given a model for rho-dependent termination of RNA chain (Fig. 10.9). Rho-dependent termination requires the Rho protein (factor) encoded by rho gene. The rho factor is a hexamer.
When a strong pause site is apparently located at specific distance from a promoter region the termination associated with rho-factor occurs. However, it must be clear that a strong pause site present close to the promoter will not act as a site for termination. Rho requires ssRNA as a binding region (Fig. 10.9 A).
Fig. 10.9 : A model for Rho factor-mediated release of RNA chain.
Rho factor moves along RNA from its binding site to the enzyme RNA polymerase. The movement is facilitated by the hydrolysis of ATP that provides energy. Hopefully, the ssRNA winds around the outside of rho factor in the large binding site. This step precedes the arrival of RNA polymerase at the termination site (B).
The RNA polymerase is partially wrapped around the outside of Rho factor. Consequently the Rho protein comes in the contact of core enzyme of polymerase. This contact is facilitated by NusA or NusG proteins. This step is dependent on the hydrolysis of nucleotide triphosphates (C).
The process of wrapping goes on continuously and disrupts the non- covalent bonds that hold the newly formed RNA to DNA and RNA polymerase. Activation of an RNA/DNA helicase activity causes the mRNA chain and core RNA polymerase to get dissociated from the DNA template. Thus, the Rho-protein-RNA complex is set free from the RNA polymerase- DNA complex (D).
Finally, core RNA polymerase is released and interacts with sigma factor that is used for the initiation of second round of transcription. A rho-dependent transcription-termination signal has also been described in bacteriophage F1 by Mose and Model (1984).
5. RNA Turnover:
On the basis of decay rates, the RNA transcribed in this way in vivo can be grouped into two: stable and unstable RNA molecules. The stable RNA molecules consist of tRNA and rRNA, whereas mRNAs are unstable. In E. coli the total population of tRNA and rRNA is 15-25% and 70-80%, respectively.
The mRNA is present only in low amount (3-5%). The reasons for the stability are association of rRNA in ribonucleoprotein complexes (ribosome) and conversion of tRNA and rRNA into the secondary structure.
This conversion protects the stable RNAs at 3′ terminus from the dissolution of ribonucleases. The cellular exonucleases degrade the mRNA in 3’→5’ direction. However, presence of stem-loop structure possibly inhibits the association of endonucleases.