Let us make an in-depth study of the polymerase chain reaction, which is a technique for amplifying DNA sequences. The below given article will help you to understand the following things:- 1. Polymerase Chain Reaction Amplifies Specific Regions of DNA 2. Performing a Polymerase Chain Reaction 3. Taq Polymerase Simplifies and Improves the Polymerase Chain Reaction and Others.
The polymerase chain reaction technique (PCR) was devised by Kary Mullis in the mid-1980s and, like DNA sequencing, has revolutionized molecular genetics by making possible a whole new approach to the study and analysis of genes. A major problem in analyzing genes is that they are rare targets in a complex genome that in mammals may contain as many as 1,00,000 genes.
Many of the techniques in molecular genetics are concerned with overcoming this problem. These techniques are very time-consuming, involving cloning and methods for detecting specific DNA sequences. The polymerase chain reaction has changed all this by enabling us to produce enormous numbers of copies of specified DNA sequence without resorting to cloning. This article describes the PCR technique and some of its novel applications.
Polymerase Chain Reaction Amplifies Specific Regions of DNA:
The PCR exploits certain features of DNA replication. DNA polymerase uses single- stranded DNA as a template for the synthesis of a complementary new strand. These single- stranded DNA templates can be produced by simply heating double-stranded DNA to temperatures near boiling. DNA polymerase also requires a small section of double-stranded DNA to initiate (“prime”) synthesis (Fig. 22.1).
Therefore the starting point for DNA synthesis can be specified by supplying an oligonucleotide primer that anneals to the template at that point. This is the first important feature of the PCR—that DNA polymerase can be directed to synthesize a specific region of DNA.
Both DNA strands can serve as templates for synthesis provided an oligonucleotide primer is supplied for each strand. For a PCR, the primers are chosen to flank the region of DNA that is to be amplified so that the newly synthesized strands of DNA, starting at each primer, extend beyond the position of the primer on the opposite strand (Figure 22.2).
Therefore new primer binding sites are generated on each newly synthesized DNA strand. The reaction mixture is again heated to separate the original and newly synthesized strands, which are then available for further cycles of primer hybridization, DNA synthesis, and strand separation.
The net result of a PCR is that by the end of n cycles, the reaction contains a theoretical maximum of 2″ double- stranded DNA molecules that are copies of the DNA sequence between the primers (Table 22.1). This is the second important feature of PCR—it results in the “amplification” of the specified region.
Performing a Polymerase Chain Reaction:
The PCR is a relatively straightforward laboratory technique, although because the technique is so versatile and the range of applications so wide, it is difficult to give a “typical” example. The starting material for a PCR is DNA that contains the sequence to be amplified.
It is not necessary to isolate the sequence to be amplified, because it is defined by the primers used in the reaction. The amount of DNA needed for a PCR is very small. In normal laboratory experiments, less than a microgram of total genomic DNA is sufficient; but, as we shall see later, the PCR can be used to amplify sequences from a single DNA molecule.
The two oligonucleotide primers directing the starting points for DNA synthesis, DNA polymerase, and a mixture of all four deoxynucleotide precursors are added to a tube containing the DNA. The total volume is usually 100 µL.
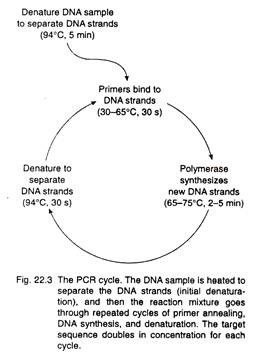
The next step in the process is to heat the reaction mixture at about 94°C for 5 minutes (Fig. 22.3). At this temperature, the double- stranded DNA molecules separate completely, forming single strands that become the templates for the primers and DNA polymerase. The temperature is then lowered to allow the oligonucleotide primers to anneal to the complementary sequences in the DNA molecules.
This annealing temperature is a key variable in determining the specificity of a PCR, so temperatures and times used vary depending on the sequences to be amplified. This generates the primed templates for DNA polymerase.
For the next step, the temperature is raised to 72°C, the optimal temperature for the heat-stable Taq DNA polymerase described in the following section. The temperature is held at 72°C for up to 5 minutes for DNA synthesis to proceed.
At the end of this period, the temperature is raised once more to 94°C, but now for only 20 seconds, so that the short stretches of double-stranded DNA (the original strand and the newly synthesized complementary strand) separate. These single strands become templates for another round of DNA synthesis, and the cycle of heating to separate strands, annealing of primers, and synthesis by DNA polymerase is repeated for as many as 30 to 60 cycles (Fig. 22.4).
Taq Polymerase Simplifies and Improves the Polymerase Chain Reaction:
Originally, E. coli DNA polymerase was used in the PCR, but this enzyme is heat- sensitive and is destroyed at the temperatures needed to separate double-stranded DNA. Therefore fresh enzyme had to be added manually for each cycle, a tedious process. An important technical advance came with the discovery that bacteria living in hot springs have DNA polymerases that work best at high temperatures.
The bacterium Thermus aquatics lives in water at a temperature of 75°C. Its DNA polymerase (Taq polymerase) has a temperature optimum of 72°C and is reasonably stable even at 94°C! Taq polymerase can be added just once at the start of a reaction and will remain active through a complete set of amplification cycles.
This development has allowed the automation of the PCR through the use of thermal cyclers, which are heating blocks that can be programmed to carry out the time and temperature cycles for a PCR. Now the Ingredients for a PCR can be placed in a thermal cycler and the reaction carried out without any manual intervention.
The specificity and sensitivity of the PCR is also improved by Taq polymerase. At the lower temperature required for E. coli DNA polymerase, the primers can anneal at sites where sequences differ slightly from the target sequences (Fig. 22.5).
Amplification can occur when mismatching primers are close together on opposite strands of DNA. Because correct complementary sequences for the primers are incorporated into the synthesized fragments, an unwanted sequence is produced with ends that precisely match the primers.
Such an “incorrect” fragment synthesized in the early cycles of a PCR will thus be efficiently amplified on subsequent cycles. In contrast, annealing of oligonucleotide primers to sites other than the desired ones is significantly reduced at the temperatures used with Taq polymerase. Consequently there is no amplification of sequences other than the targeted sequence.
The specificity is improved further in the “hot-start” method, where all the reagents save one are heated to 72°C before adding the final ingredient, for example, the Taq polymerase. This increase in specificity simplifies the analysis of PCR products. The amplified target fragment can be seen easily on an ethidium bromide-stained gel because the background staining of non-target sequences is eliminated.
Fidelity of DNA Synthesis by Tag Polymerase Determines the Accuracy of Polymerase Chain Reaction Amplification:
Like all other biochemical processes, DNA replication is not a perfect process, and occasionally DNA polymerase will add an incorrect nucleotide to the growing DNA chain. The rate of misincorporation measured in a naturally replicated DNA molecule is approximately 1 in 109 nucleotides. Cells achieve such extraordinary accuracy because the DNA replication machinery removes mismatched nucleotides added to the DNA chain.
In vitro Taq polymerase docs not have this “proofreading” capability, and using the temperatures and salt concentrations typical of a PCR, the enzyme incorporates one incorrect nucleotide for about every 2×104 nucleotides incorporated.
This is not a serious matter for bulk analysis of PCR products because molecules with the same misincorporated nucleotide will form a very small portion of the total number of molecules synthesized. But misincorporation is important if PCR fragments are to be used for cloning (Fig. 22.6).
Each clone is derived from a single amplified molecule. If this molecule contains one or more misincorporated nucleotides, then all the cloned DNA in that clone will carry the identical “mutation”. This problem can be reduced by beginning the PCR with a large, rather than a small, number of template molecules. Fewer cycles of amplification are needed, and less total DNA synthesis takes place.
DNA for the Polymerase Chain Reaction Comes from a Variety of Sources:
DNA for a PCR is often total genomic DNA extracted from cells. However, the PCR does not require highly purified DNA, and DNA released by the boiling of cells can be used directly without any purification. The PCR can also be used to study the pattern of gene expression: mRNA is converted to cDNA using reverse transcriptase, and the cDNA then serves as the template for the PCR. DNA sequences do not have to be isolated before amplification by a PCR, because the specificity of the reaction is determined by the oligonucleotide primers.
For example, a PCR is a very convenient way of preparing DNA from cloned inserts in plasmid or bacteriophage vectors (Fig. 22.7). All that is needed are oligonucleotide primers complementary to the vector sequences on either side of the cloning site. Any insert is easily amplified in this way regardless of its sequence. This PCR method is now being used routinely to screen for clones containing inserts.
DNA is a very stable molecule, provided it is not exposed to nucleases, and DNA from the most extraordinary sources has been used for PCRs. For example, human papilloma virus DNA has been detected in cervical carcinoma biopsies embedded in paraffin for over 40 years. Human archival materials that can provide DNA for PCR are not restricted just to those embedded in paraffin. Blood samples for the neonatal, detection of phenylketonuria have routinely been taken by heel-prick of newborns arid stored as dried spots on cards for many years.
As much as 1 µg of DNA can be extracted from a blood spot; this is more than enough to be used for PCR analyses of human disease genes such as the genes for phenylketonuria and cystic fibrosis. As we shall see later, even DNA isolated from Egyptian mummies several thousand years old has been amplified by the PCR.
Polymerase Chain Reaction is used to Amplify Human-Specific DNA Sequences:
When human cells are fused with rodent cells, the human chromosomes are preferentially lost from the hybrid cells, so that hybrid cell lines containing only one human chromosome—or even a part of a human chromosome, can be produced. These cell lines are a convenient source of DNA for cloning if the fragment of chromosome carried by the cells contains the desired gene.
So-called Alu-PCR provides a very simple means of characterizing and amplifying just the human DNA in these hybrid lines. The method uses primers for repetitive sequences that are inserted at many places in genomes. One of these elements, the Alu repeat, is present in as many as 9, 00,000 copies in the human genome.
This 300-bp Alu repeat is very variable, but it contains a sequence that is human-specific. Two oligonucleotide primers were made, one in each direction, using the most conserved section of this sequence (Fig. 22.8). The primers cannot be used together because they hybridize to each other.
However, Alu sequences can be present in either direction in DNA, so the primers can be used individually, and human sequences will be amplified if they lie between adjacent Alu repeats that are pointing in opposite directions (Fig. 22.8). Varying numbers of human DNA fragments are produced depending on the size of the human DNA in the cell line. This technique is widely used for “rescuing” human DNA sequences from other DNAs.
Polymerase Chain Reaction Products can be Sequenced Directly:
Like any DNA, the double-stranded products of a PCR can be sequenced. However, because single-stranded DNA is the best template for Sanger’s dideoxy chain termination method, a technique called asymmetric PCR has been devised to produce single-stranded DNA (Fig. 22.9).
The standard PCR is set up except that the concentration of the two primers differs by a factor of 100. Double- stranded DNA fragments are produced until the limiting primer is used up. The remaining primer continues to anneal and prime DNA synthesis, producing only one of the two strands. Although this strand accumulates at a linear rather than an exponential rate, sufficient single-stranded DNA is produced for sequencing.
Detecting Mutations using Polymerase Chain Reaction Amplification:
Mutations occur in cancers and in inherited disorders. Knowing the nature of the mutation in a patient is important for diagnosis and therapy. PCR amplification is proving invaluable as a tool for screening particular genes for mutations. As we shall see later in this chapter, the PCR can be used to follow the fate of cancer cells after therapy; and in we shall discuss examples of the increasing number of human inherited disorders being diagnosed in this way.
In 1989, Harold Varmus and J. Michael Bishop were awarded the Nobel Prize for their discovery that the oncogenes carried by RNA tumor viruses are part of the normal genetic makeup of the cell; cancers arise when these normal genes mutate.
This major advance in cancer research has been taken further with the recognition that certain cancers are caused by specific and reproducible mutations. For example, mutations in the ras oncogene have been recognized in many human cancers, and the PCR has been used to analyze the pattern and frequency of mutations in the human RAS genes. (By convention, human oncogenes are written in capital letters.) Samples from large numbers of patients were screened rapidly by the PCR, and the studies revealed that different forms of lymphoid malignancies had different RAS mutations.
Genetic analysis has important practical implications for retinoblastoma, a childhood cancer of the eye caused by mutations in a gene at q14 on chromosome 13 (“q” refers to the long “arm” of the chromosome; “14” refers to a particular location on this arm). There is a heritable form of retinoblastoma in which there is a germ line mutation at one of the two retinoblastoma alleles.
All the cells of these patients contain one normal and one mutant allele, and a single mutation at the normal allele is all that is required to develop the cancer. Retinoblastoma can also arise spontaneously in patients who have not inherited a mutant allele if a retinal cell suffers two mutations, one in each allele.
Thus in families with just one affected member, it has been difficult to determine if the hereditary form of retinoblastoma is involved. This can now be done by using the PCR and sequencing to analyze mutations in tumor tissue and normal tissue from affected individuals. Mutations in some patients are found only in tumor tissue, suggesting that these are spontaneous mutations and that the children of these patients will not be predisposed to developing retinoblastoma.
In other cases, the mutation is found in normal tissue as well as in the tumor cells, showing that these patients are born with one mutation in all their cells. The children of these patients will be at a greatly increased risk of developing retinoblastoma because they may inherit a mutated copy of the retinoblastoma gene. These findings are important for genetic counseling.
Polymerase Chain Reaction Amplification is used for Monitoring Cancer Therapy:
The ability to detect genetic lesions characteristic of tumor cells is a valuable tool for the oncologist trying to determine if a patient being treated for a leukemia is free of malignant cells. The physician would like to stop treatment with cytotoxic drugs or radiation as soon as the cancer is destroyed, and conversely would want to resume treatment as soon as a relapse begins.
As we shall learn in, some cancers arise as a result of chromosomal translocations that involve known genes. For example, there is a translocation between chromosomes 14 and 18 in follicular lymphomas. With such mutations as markers, these cancer cells can be detected by conventional Southern blotting if they are present at concentrations of 1 in 100 normal cells, so that a patient judged cancer- free by Southern blotting may still harbor significant numbers of cancer cells.
In contrast, PCR techniques are capable of detecting as few as 1 cancer cell in 106 normal cells, providing a much more sensitive indicator for the oncologist. The two PCR primers are chosen from the sequences adjacent to the breakpoints on each chromosome. It is only in cells with the translocation that the primers are brought together so that the sequences between them can be amplified (Fig. 22.10).
A similar strategy has been devised for detecting leukemia’s using mRNA as the starting material. This has the advantage that mRNA already represents an amplification over genomic DNA in that a cell contains many copies of the mRNA for a gene.
Polymerase Chain Reaction Amplification is used to Detect Bacterial and Viral Infections:
Similarly, PCR techniques can be used to monitor bacterial or viral infections. Conventional diagnostic procedures are based on the ability to grow organisms in culture or to detect their presence in patients by using antibodies. One difficulty with these types of tests is that the former may require several weeks before diagnosis is possible and the alternatively insensitive.
These are particularly important considerations for diagnosis of AIDS or for studies of the epidemiology of human, immunodeficiency virus (HIV) infections. Here, as in cancer therapy, the aim is to detect rate infected cells among a large population of uninfected cells. To detect HIV, PCR primers are made for sequences in the virus, and a PCR is carried out using DNA extracted from peripheral blood cells.
This approach detects integrated HIV DNA in infected cells. The presence of viral RNA is thought to indicate an active infection, and this can be diagnosed by performing PCR using cDNA templates produced by reverse transcription of RNA from infected cells.
Tuberculosis is caused by Mycobacterium tuberculosis; diagnosis is difficult because there may be too few organisms in pathological samples for histological diagnosis. Instead, the pathogen has to be identified following growth in culture and antibiotic sensitivity testing, a procedure that takes several weeks.
This is clearly a situation in which PCR technique should help. PCR amplification has been performed using primers for a sequence within a gene that is highly conserved in all mycobacterial species. The amplified DNA fragment was hybridized with species-specific probes to identify the specific strain involved.
The PCR-based test proved far more rapid than the conventional test and far more sensitive, detecting as few as 10 bacilli in 106 eukaryotic cells. It is clear that PCR techniques will soon be adapted to routine diagnostic use in clinical microbiology laboratories.
Polymerase Chain Reaction Amplification is used for Sex Determination of Prenatal Cells:
One area in which genetic analysis of a small number of cells is very important is in prenatal diagnosis. For inherited X-linked disorders that affect only males, sex determination is the first step in prenatal diagnosis. Male sex determination using DNA is possible because males carry unique sequences on the Y chromosome.
Some of these are repeat sequences, already “amplified” relative to normal genes. For example, the 3.5-kb DYZI sequence is present on the Y chromosome in as many as 5,000 copies. PCR techniques can be used to amplify a 149-bp fragment from the DYZI sequence, specific to males.
In one experiment to perform prenatal sex determination after in vitro fertilization, researchers removed a single cell from each 10-cell human pre-implantation embryo using a micromanipulator, and the DYZI sequence was amplified from each cell using 60 cycles of the PCR (Fig. 22.11). An amplified fragment was obtained only from cells from male embryos.
This procedure has now been used clinically for families at risk for X-linked inherited disorders, with implantation of biopsied embryos into mothers. The speed of PCR allows biopsy, sex determination, and transfer of the embryos to the mothers in the same day.
Usually there are sufficient numbers of female embryos for two embryos to be transferred, and their subsequent progress is monitored by assay of chorionic gonadotrophin levels. The sex of these fetuses is checked by karyological analysis of chorionic villus cells. There are now five apparently normal, healthy girls born following this procedure.
The PCR can amplify repeated sequences from single embryonic cells, but can it be used to amplify single genes important in human inherited diseases? This question has not yet been answered directly. Instead, unfertilized human oocytes were used as equivalents for single embryonic cells.
Sequences from two genes those for Duchenne muscular dystrophy and cystic fibrosis—were amplified from these single cells, and the appropriate fragments were detected. In order to detect an amplified fragment from the cystic fibrosis gene, amplification had to be carried out for 80 cycles.
It is possible to determine that a fetus is male by cytological detection of male cells in the peripheral blood of a mother carrying a male fetus. However, the reliability of this procedure is poor and precludes its use for prenatal sex determination. It has been estimated that the number of fetal male cells present in the maternal circulation is less than 1 per 70,000 maternal cells, but, as we have seen, PCR techniques are capable of detecting such rare cells.
In one study, blood samples were taken from pregnant women as early as after 9 weeks of gestation, DNA extracted, and part of the DYZ1 sequence amplified. After 40 cycles of amplification, an aliquot of the reaction mixture was used for a further 15 to 20 cycles of amplification.
The primers used for this second round-of amplification were internal to the first pair of primers. So-called nested primers reduce significantly the chance of amplifying unwanted sequences when a large number of amplification cycles are used (Fig. 22.12). Sexes of all fetuses in this study were determined correctly.
Fetal sampling, whether by chorionic villus biopsy or by amniocentesis, carries a small but significant risk of miscarriage, so the advantage of using maternal blood for fetal sexing is clear. Whether this technique can be developed for routine use is another matter. Quite extraordinary lengths had to be taken to prevent contamination of samples by male DNA (see below).
Polymerase Chain Reaction Methods Permit Linkage Analysis Using Single Sperm Cells:
Determining recombination between genes at meiosis has been the classical way to map genes ever since its application to Drosophila by A. H. Sturtevant and T. H. Morgan. This is difficult for mapping human genes because linkage studies have to be performed in families, where mattings cannot be controlled and the number of offspring is low (see Arnheim et al for a further discussion of human gene mapping). PCR analysis of alleles in sperm offers a way of performing linkage analysis with single cells.
A chromosome contained in a sperm is a single meiotic product, and by examining two loci at a time and determining the frequency of recombination between them, it should be possible to derive the genetic distance between the loci.
For the geneticist, examining 1,000 sperms is like studying a family with 1,000 children. These estimates of genetic distances are for male chromosomes, an important point, given that recombination frequencies for some genes differ for male and female meioses.
A preliminary experiment using loci with a known recombination rate was performed to determine the feasibility of this approach. The loci chosen—the parathyroid hormone gene locus (PTH) and the Gϒ globin locus (HBG2) on the short arm of human chromosome 11— have a recombination frequency of about 0.15 based on family analysis.
For this PCR experiment a total of 708 sperm from two donors heterozygous at both loci were analyzed. The genes were amplified and then tested with allele-specific oligonucleotide probes to detect the four possible meiotic products (Fig. 22.13). To calculate a recombination frequency, technical complications have to be taken into account; for example, two sperm may be in the same tube.
As a consequence, a complex statistical treatment of the data is necessary. Nevertheless, the frequency of recombination as calculated in this experiment was 0.16, in very good agreement with the estimate from family studies.
The combination of PCR and allele-specific oligonucleotide probes offers a promising method for fine-resolution mapping of human genes. One current limitation is isolating sufficient numbers of single sperm cells for analysis, but this may be overcome by using fluorescence-activated cell sorters.
Polymerase Chain Reaction Techniques are used in Studies of Molecular Evolution:
Molecular genetic information is being used increasingly in evolutionary studies to determine the degree of relatedness between species and, in so doing, to build up family trees similar to those produced by classical comparative methods. It is assumed that as species diverge from a common ancestor, their nucleotide sequences diverge and that measurements of the degree of nucleotide divergence (or, conversely, of nucleotide homology) can be used to determine relationships.
Homology can be measured by determining the degree of hybridization between total DNA from Individuals, by comparing nucleotide changes in the same gene between species, or by comparing mitochondrial genes. Mitochondrial genes have the advantage that the mitochondrial genome is not rearranged during meiosis and they have high point-mutation rates so that changes can be measured over shorter time periods.
The usefulness of molecular methods has been limited by the necessity of working with living species that can provide DNA samples. Thus, relationships between living organisms can be examined directly, but the relationships of living organisms to extinct organisms can only be inferred.
However, tissue samples from extinct and rare species preserved in museums throughout the world are a vast resource, and DNA has been Isolated from sources as varied as museum skins, human mummies, dried plants, and even soft tissue specimens in preservatives.
Unfortunately, the DNA molecules are very short because of degradation; they are modified because of damage from environmental mutagens such as ultraviolet radiation; and they may be heavily contaminated with bacterial DNA. Such DNA is not suitable for studying by conventional cloning techniques. For example, the average size of DNA fragments cloned from 4000- year-old human mummy tissue was only 90 bp.
The PCR has changed the situation dramatically. The PCR efficiently amplifies small fragments of DNA, such as are more likely to remain intact in ancient samples of DNA, and PCR methods can amplify these intact molecules even if very few are present. Remarkably, it seems that Taq polymerase can synthesize full-length target sequences, even if copies of the target sequence are fragmented, provided these fragments overlap.
As the amplified fragments Increase in concentration, two different fragments can anneal to each other by their overlapping sequences, and function as primers. Each fragment is therefore extended using the other fragment as a template, so that even in this situation a complete target sequence is synthesized.
For example, analysis of mitochondrial DNA has been used similarly to elucidate the phylogenetic position of Thylacinus cynocephalus, the extinct marsupial wolf. Sequences from the mitochondrial cytochorome b and 12S ribosomal RNA genes were amplified using DNA isolated from the hides of museum specimens and compared with the same sequences from living marsupials and the South American opossum.
Sequence analysis of the PCR products showed clearly that Thylacinus cynocephalus is related more closely to marsupials and justifies its common name of the marsupial wolf. The PCR has even been used to amplify DNA sequences from fossils. These fossils may be formed under conditions that promote a remarkable degree of preservation. Leaves in shale fossil beds dating from the Miocene Period (18 million years ago) are so well preserved that the leaves are still green.
DNA has been extracted from fossilized leaves scraped off the rocks and used for a PCR with primers for part of the ribulose 1,5-biphosphate carboxylase gene (rbcL). Astonishingly, the correct size of fragment, 820 bp long, was produced—from DNA molecules 18 million years old!
Furthermore, asymmetric PCR was used to produce single-stranded molecules for sequencing. Comparisons of the rbcL sequence of the fossil plant with that of existing species identified this extinct Miocene plant as a member of the Magnolia family.
PCR has also been used in studies of the evolution of extant human populations. For example, mitochondrial sequences in DNA from freshly plucked hair were amplified using the PCR and analyzed to determine the genealogical relationships of the Kung people. These are aboriginal southern Africans who speak click language and who have remained genetically isolated. This study supported the notion of the African origin of human mitochondrial DNA.
Contamination can be a Problem in Polymerase Chain Reaction Studies:
An unforeseen and unwelcome corollary of the amplification power of the PCR is that minor contamination of the starting material can have serious consequences. For example, in the study of prenatal sex determination using maternal blood samples described previously, there was one positive sample from a non-pregnant negative control.
This was attributed to contamination by minute amounts of tissue shed from the skin of a male worker. It proved necessary to have only women preparing and analyzing the DNA samples. In another case, DNA from one of the museum specimens of the marsupial wolf yielded human mitochondrial sequences when amplified.
In this case, the contaminating PCR product was readily identified as a contaminant by sequence analysis, but the contamination of ancient human DNA by contemporary human DNA might be much more difficult to detect. Contamination may prove to be a significant problem in forensic applications of the PCR. Biological samples associated with crime scenes are rarely in the pristine state of the materials used to prepare DNA in the laboratory.
One common source of contamination is the products of previous amplification reactions. A completed PCR reaction mixture may contain as many as 1013 amplified fragments, so that even minute volumes such as droplets in an aerosol from a pipette tip contain very large numbers of amplifiable molecules.
One solution to contamination problems is to physically separate pre- and post-amplification steps. A clever development uses uridine triphosphate (UTP) instead of deoxythmidine and includes the enzyme uracil-N-glycosylase (UNG) in the PCR reaction mixture. UNG will degrade any DNA contamination from a former PCR because the previously amplified DNA contains UTP.
UNG is then inactivated by the high temperature of the new PCR, so the products of the new reaction can accumulate. It is always essential to run negative controls that will reveal the presence of contaminating DNA in the reaction reagents in every PCR experiment.
The Polymerase Chain Reaction—A Technical Revolution in Molecular Genetics:
The versatility of the polymerase chain reaction is enormous, and the combination of the PCR and sequencing is an extraordinarily powerful tool for the analysis of genes. The most spectacular demonstration of this is the molecular identification of the Magnolia plant from the Miocene Period. Who could have imagined that it would ever be possible to analyze in detail genes from DNA 18 million years old? Other applications of the PCR described later in this book are no less remarkable.
For example, it is used for cloning genes, for in vitro mutagenesis, in mapping and sequencing large genomes, and in diagnosing human inherited disorders. In 1985 there were but three research reports on the PCR; 5 years later it is being used in thousands of laboratories. The PCR has indeed revolutionized the practice of molecular genetics.