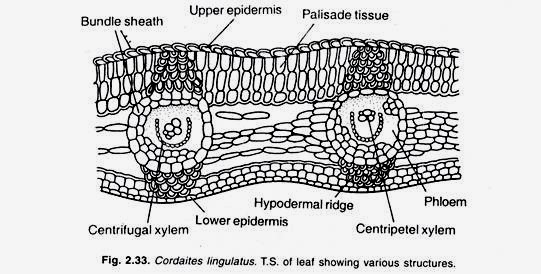
As might be expected, the regulation of enzyme production in eukaryotic cells is much more complex than in prokaryotic cells.
Eukaryotic cells contain a number of chromosomes instead of the single chromosome found in prokaryotes. Moreover, eukaryotic cell chromosomes are diploid and at times polyploid.
The DNA of the chromosomes is usually supercoiled and highly folded and the chromosomes themselves are physically separated from the cytoplasmic ribosomes by the nuclear envelope.
Undoubtedly, these factors make control of enzyme synthesis more complex, even though transcription and translation involve what is basically the same mechanism as in prokaryotes.
However, structural genes for a group of functionally related enzymes that might constitute an inducible component such as an operon are not found adjacent to one another on a chromosome and may, in fact, be distributed among different chromosomes.
Enzyme induction does occur in primitive eukaryotic organisms such as yeast and Neurospora, but operons are either few or nonexistent in higher eukaryotes. In all but a few cases, the mRNAs of higher eukaryotes contain the coding sequence of only one structural gene (i.e., they are monogenic).
The induction process in the fungi is slower, and the change in concentration of enzymes is not as great as in the prokaryotes. In yeast, the induction of β-galactosidase takes much longer than in E. coli; also, the increase in enzyme activity is 10-fold and not a 1000-fold. The enzyme tryptophan-2,3-oxygenase found in the liver cells of vertebrates is inducible but induction requires many hours.
1. Z-DNA:
In addition to the B form of the DNA double helix, there is a Z form. In Z-DNA, the double helix is left-handed, each polynucleotide containing sequences of alternating purines and pyrimidines. A number of observations suggest that Z-DNA may play a role in the regulation of gene expression. For example, Z-DNA is formed in the transcriptionally active macronucleus of Stylonychia mytilis (a protozoan) and in the interband regions of Drosophila salivary chromosomes. Proteins associated with polytene chromosomes of Drosophila bind antibodies specific for Z-DNA. Some regulatory proteins appear to bind to Z-DNA but not B-DNA, including the catabolite activator protein described earlier.
2. Calcium Ions and Calmodulin:
In the past few years, it has become quite clear that calcium ions play a very important role in the regulation of certain activities in eukaryotic cells. For example, such diverse phenomena as cell motility, muscle cell contraction, chromosome movement, endocytosis, exocytosis, cyclic nucleotide metabolism, phosphorylation of proteins, and glycogen metabolism are influenced by the level of Ca2+ in the cytosol. Some of the effects of calcium ions are mediated through a specific calcium-binding protein called calmodulin (Fig. 11- 16) found in nearly all cells.
Calmodulin is a small protein (its molecular weight is 16,720) consisting of a single polypeptide chain containing 149 amino acids. Especially interesting is the finding that its primary structure is essentially the same in all of the species that have been studied so far, indicating that the protein has undergone little change in the course of evolution.
Each molecule of calmodulin binds four calcium ions and its common occurrence in eukaryotes suggests that it has a universal regulatory function along with Ca2 +. When Ca2+ enters the cytosol, it is bound by calmodulin to form a complex that then influences cellular activity by interacting with certain other proteins. Some of the proteins affected by the Ca2 + – calmodulin complex are structural proteins but most appear to be enzymes.
For example, the Ca2 +-calmodulin complex serves to directly activate a family of enzymes called protein kinases. Which protein kinases are activated may depend on the amount of Ca2+ in the cytosol. This is because calmodulin can bind up to four calcium ions and a complex containing only one Ca2 + may activate a different enzyme than a complex containing two Ca2+ (and so on). The effect of the Ca2+ – calmodulin complex on cell metabolism may be indirect.
For example, the Ca2 +-calmodulin complex acts to trigger adenylate cyclase activity in the plasma membranes of cells and these results in the production of cAMP. cAMP’s role as a “second messenger” influencing cell metabolism is discussed below. Additional examples of the manner in which calmodulin and Ca2 + regulate cell metabolism are given in Table 11-4.
3. Enzyme Induction by Hormones:
Enzyme induction in prokaryotic cells is usually triggered by a potential metabolite (such as the induction of β-galactosidase by lactose). As noted above, this form of enzyme induction occurs in some eukaryotic cells as well; however, in higher animals there also is a highly developed control system in which hormones act as messengers that coordinate the biosynthetic activities of cells.
This coordination may take the form of activating (or deactivating) enzymes that already exist in the cell or the effect may be on gene expression, resulting in the production of additional enzymes. The term “messenger” is appropriate, because the hormones are produced and secreted by one cell (or tissue) and have an effect on a different cell (or tissue).
The two tissues producing and responding to the messenger may be in widely separated parts of the body, the messenger traveling from one site to the other via the bloodstream. Sometimes, the effect of the messenger is to cause the “target” cell to produce and secrete a different hormone (Fig. 11-17a).
From a chemical point of view, hormones are quite diverse. Some hormones (e.g., thyroxin and epinephrine) are small molecules derived from amino acids and others are proteins (e.g., insulin and erythropoietin) or steroids (e.g., progesterone and Cortisol). Hormones regulate the production of enzymes through a variety of mechanisms. For example, the hormone may be the first messenger among a series of messengers that regulate a metabolic response (Figs. 11-17b and 11-17c). Such hormones attach to receptor sites on the plasma membrane of the “target” cell. The receptor sites consist of specific proteins having high affinities for the hormones.
Hormone binding at a receptor site serves to initiate a response by the cell. Different tissues may be affected by the same hormone. The cells in these tissues may contain similar hormone-binding receptors, but the interaction between receptor and hormone is followed by a different cellular response.
Second Messengers:
In some cases it appears that the hormone receptor of the target cell is associated with an enzyme in the plasma membrane. Hormone binding by the receptor changes the receptor-enzyme complex and activates the enzyme. Once activated, the enzyme catalyzes a reaction that forms a second messenger that passes into the cytosol and triggers further responses in the target cell.
The most common second messenger is cyclic AMP. It is formed by the enzyme adenylate cyclase, a membrane-bound enzyme associated with hormone receptor sites. cAMP produced by adenylate cyclase acts as a second messenger in metabolic pathways associated with the breakdown of glycogen and triglycerides (Fig. 11-18). cAMP appears also to be involved in cellular reactions that produce and secrete hormones.
The cellular response induced when cAMP acts as a second messenger continues for only a brief period because of the presence of phosphodiesterases in the cytosol. These enzymes degrade cAMP, producing the inactive, qoncylic mononucleotide 5′-AMP. Calcium ions also act as second messengers. Some surface receptors are associated with channels through the plasma membrane. When the receptor binds the first messenger (i.e., the hormone) the channel is opened temporarily and Ca2+ enters the cell. Once inside the cell, the Ca2+ is bound by specific proteins such as calmodulin.
The activation of enzymes by calmodulin was described earlier. It turns i out that many of the enzymes activated by cAMP are also activated by Ca2 +-calmodulin and Ca2 + – calmodulin affects the activities of enzymes that form and degrade cAMP. In turn, cAMP influences the plasma membrane channels through which Ca2 + passes by phosphorylating the proteins that line these channels.
Cyclic GMP (cGMP), like cAMP, activates certain protein kinases. However, cGMP does not appear in response to the binding of hormones by plasma membrane receptors. Rather, the level and activity of cGMP appear to increase as the intracellular level of free Ca2 + increases.
“Double” Second Messengers:
Many hormone receptors are associated with a group of plasma membrane phospholipids called polyphosphoinositides. Binding of the first messenger (hormone) to the receptor initiates the breakdown of the membrane-bound polyphosphoinositides, thereby forming diacylglycerol and inositol triphosphate.
The diacylglycerol enters the cytosol where it acts as a second messenger, activating a protein kinase (Fig. 11-19). (The protein kinase activated by diacylglycerol has different properties than the protein kinase activated by cAMP and Ca2 + -calmodulin.) The inositol triphosphate produced in the plasma membrane also enters the cytosol and acts as a second messenger. When inositol triphosphate reaches the intracellular membranes, it induces the release of Ca2 +. Ca2 + released into the cytosol then activates certain proteins such as calmodulin.
4. Effects of Hormones on Gene Expression:
The binding of certain hormones to plasma membrane receptors is followed by internalization. Internalization takes the form of an infolding of that portion of the plasma membrane containing the receptor- hormone complex, thereby forming a vesicle.
The vesicle subsequently fuses with a Golgi body or lysosome, the enzymes of which presumably alter the receptor-hormone complex. Ultimately, a product is formed that enters the cell nucleus where it interacts with the genome, thus altering gene expression.
Steroid hormones (e.g., estrogen and progesterone) (first messenger) enter the target cell by passing directly through the plasma membrane and into the cytosol. Within the cytosol, the hormones combine with receptor molecules and the complexes then migrate to the cell nucleus, where they have a direct impact on the expression of certain genes (Fig. 11-20).
In vitro studies using chick oviduct cells have shown that the hormone estrogen enters the cytosol to form a hormone-receptor complex that migrates to the cell nucleus and induces the increased rate of transcription of the genes for lysozyme and ovalbumin.
5. Protein Phosphorylation and Metabolic Control:
Phosphorylation by protein kinases appears to be one of the major mechanisms for activating and inactivating a great number of enzymes and thereby controlling many processes in eukaryotic cells. Generally speaking, many enzymes in degradative (catabolic) pathways are activated by phosphorylation, whereas the enzymes in synthetic (anabolic) pathways are inactivated by phosphorylation.
Protein kinases activated by Ca2 + -calmodulin, cAMP, and diacylglycerol phosphorylate key enzymes of glycolysis and other bio- degradative pathways and thereby activate these pathways. The protein kinases also phosphorylate key enzymes in glycogen metabolism, gluconeogenesis, fatty acid synthesis, cholesterol synthesis, and protein synthesis and thus inactivate these pathways. Protein kinases catalyze protein phosphorylation using ATP as the source of phosphate.
Protein phosphatases are enzymes that catalyze the removal of phosphate from phosphorylated enzymes (or other proteins). It has been proposed that protein phosphatases also function in metabolic control because they can dephosphorylate glycolytic enzymes and thereby inactivate them and can dephosphorylate glycogen-synthesizing enzymes and thereby activate them.
Although the evidence is not yet available, it is likely that protein phosphatases dephosphorylate the enzymes of gluconeogenesis, fatty acid synthesis, cholesterol synthesis, and protein synthesis and therefore activate these enzymes as well.
6. Amplification of Signals:
Signals to cells in the form of a few molecules of a hormone or other first messenger are greatly enhanced by mechanisms that use second messengers, double messengers, or even third messengers. Although only a few molecules of the first messenger may bind to the surface receptors, they activate enzymes each of which produces many copies of the second messenger. If additional messengers are produced by enzymes activated by second messengers, thousands of signals will be produced in the cytosol within a very short time.
7. Repression of the Genome in Eukaryotes:
Unlike prokaryotic cells, the cells of higher animals and plants undergo extensive differentiation, forming ‘ tissues that have specific and limited physiological roles. Yet most differentiated cells of higher plants and animals contain complete genomes.
This implies that large segments of the genome are repressed and go unexpressed. RNA-DNA saturation hybridization experiments indicate that only 6- 30% of the genome is expressed. In some cases, the repression is reversible.
For example, when differentiated carrot root cells are isolated and grown in tissue culture, new differentiated cells are produced, including cells that are characteristic of vascular tissue, storage tissue, and epidermal tissue. However, in many differentiated cells, such as nerve and muscle cells of higher animals, large portions of the genome are permanently repressed.
The mechanism of gene repression of eukaryotic cells is not yet clear. The chromosomes of eukaryotic cells contain large quantities of proteins and RNA in addition to DNA (Table 11-5), and over the years these have been considered potential repressors. However, their chemical properties do not adequately support such a contention.
For example, Stedman and Stedman proposed as long ago as the 1940s that the histones might act as gene repressors. However, there are only five major classes of histones in eukaryotic cells and they occur in about equal amounts, with little variation between different tissues of an organism or between species.
It would therefore appear that his- tone function is more fundamental. If the histones do have repressor activity, then the effect is nonspecific. Electrophoretic analysis of the non-histone proteins in chromatin reveals that they occur in much greater variety than the histones. However, their diversity is insufficient to support the contention that they play a role in gene repression.
8. The Britten-Davidson Model of Gene Regulation in Eukaryotes:
Although several models have been proposed to explain gene regulation in eukaryotes, none has been documented with evidence that would give it the degree of certainty that surrounds the Jacob-Monod operon model for prokaryotic cells. However, the model proposed by R. Britten and E. Davidson in 1969 has attracted a great deal of attention.
According to this model (Fig. 11-21), the nuclear chromosomes contain DNA sequences called sensor genes that recognize various cellular substances such as metabolic inducers (substrates), hormone-receptor complexes, or regulatory nucleotides (e.g., ppGpp).
When the inducer enters the nucleus, it binds to the sensor and promotes the transcription of an adjacent integrator gene whose product is a specific activator RNA. The activator RNA can attach to appropriate DNA sequences that constitute receptor sites on either the same or a different chromosome. Presumably, the function of the activator would be analogous to that of the cAMP-CAP for prokaryotes.
The binding of the activator to a receptor site promotes transcription of adjacent structural genes. After undergoing some modification called “processing” the mRNA transcript leaves the nucleus and is translated into protein.
A number of elaborate modifications of the basic model can explain the variations in gene expression and differentiation in eukaryotes. For example (Fig. 11-22a), a number of different sensor genes (S1, S2, and S3) on binding different inducers (I1, I2, and I3) promote the formation of different activator RNA molecules (a, b, and c) by their companion integrator genes.
The presence of multiple receptor sites for each structural gene (i.e., L, M, and N) would imply that different combinations of structural genes would be transcribed, depending on binding of the various activators to their respective receptor sites. The binding of an activator to any one receptor would trigger transcription of the adjacent structural gene.
In an alternative model (Fig. 11-22b), transcription of structural genes in various combinations results from the binding of a specific inducer. In this variation, the sensors initiate activator synthesis in a number of adjacent integrator genes, and each activator then associates with one receptor.
The models have a number of interesting possibilities and are supported by the observation that a large proportion of the DNA in eukaryotic cells (e.g., 40% in calf thymus gland cells) consists of repeated nucleotide sequences that are too small to be structural genes; these, perhaps, are receptor sequences.
9. Compartmentalization:
A final regulatory mechanism in eukaryotic cells is the physical separation and isolation of groups of enzymes within membranous boundaries, that is, specific groups of enzymes are compartmentalized within the cellular organelles. For example, in both plant and animal cells the enzymes of the tricarboxylic acid or Krebs cycle are physically separated from those of glycolysis because the former are confined within mitochondria and the latter are present in the cytosol.
In plant cells, the enzymes of the dark reactions of photosynthesis are physically isolated within chloroplasts. The enzymes of the glyoxylate cycle are compartmentalized in micro- bodies, whereas many of the cell’s powerful hydrolytic enzymes are restricted within the lysosomes.
Many of these compartmentalized enzymes act on substrates or employ cofactors that are produced by enzymes that are restricted to other parts of the cell. Regulation of the transport of these compounds across cellular membranes from one cell compartment to another affords yet another level at which the control at metabolsim can be exercised.