Read this article to learn about Composition and Structure of the Nucleic Acids: DNA & RNA !
There are two major classes of nucleic acids: DNA and RNA.
Both are composed of un-branched chains of units called nucleotides, each of which contains:
(1) A nitrogenous base (either a purine or pyrimidine),
(2) A pentose, and
(3) Phosphoric acid.
In RNA, the pentose is ribose, whereas in DNA it is 2-deoxyribose. Both DNA and RNA contain the purine nitrogenous bases adenine (abbreviated A) and guanine (G) and the pyrimidine cytosine (C), but in DNA a second pyrimidine is thymine (T), whereas in RNA it is uracil (U).
A number of other nitrogenous bases have been identified in DNA and RNA, but these occur much less frequently. The phosphoric acid component of each nucleotide is, of course, chemically identical in both nucleic acids. These relationships are summarized in Table 7-1, and the corresponding chemical formulas are shown in Figure 7-3.
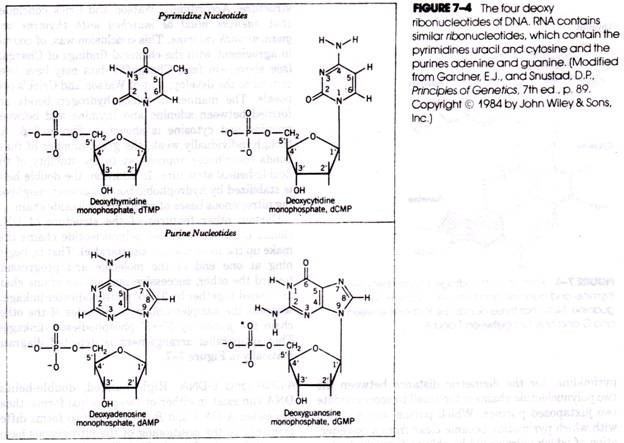
The pentose of each nucleotide unit is simultaneously bonded through its number 1 carbon atom to the nitrogenous base (forming a nucleoside) and through its number 5 carbon atom to phosphoric acid. The structures of the four deoxyribonucleotides of DNA together with the specific numbering system used to identify each constituent atom are shown in Figure 7-4. Successive nucleotides of DNA and RNA are joined together by phosphodiester linkages involving the 5′-phosphate group of one nucleotide unit and the 3′-hvdroxyl group of the neighboring unit (Figure 7-5).
The “backbone” of a nucleic acid molecule is formed by the repeating sequence of pentose and phosphate groups, and this is the same in all molecules. What distinguishes one DNA (or RNA) molecule from another is the specific sequence of purine and pyrimidine bases present in the chain of nucleotides and the total number of nucleotides (i.e., the size of the molecule). Each chain of nucleotides is called a polynucleotide.
Structure of DNA:
At one time, it was believed that the four purines and pyrimidine’s of DNA occurred in approximately equal amounts in the molecule. However, the studies of E, Chargaff and others in the late 1940s showed that this was not the case. Instead, they found that the relative amounts of the nitrogenous bases varied between species but were constant within a species.
The constancy noted within a species was maintained regardless of the tissue or organ from which the DNA was isolated. Furthermore, the relative amounts of the nitrogenous bases were similar in closely related species and quite different in unrelated species.
Chargaff also made the following extremely important finding. Regardless of the species used as the source of DNA, the molar ratios of adenine and thymine were always very close to unity, and the same was true for guanine and cytosine. No such constant relationship could be demonstrated for any other combination of nitrogenous base pairs. This implied that for some reason, every molecule of DNA contained equal amounts of adenine and thymine and also equal amounts of guanine and cytosine.
Using chemical information of this sort, together with the results of X-ray crystallographic studies of DNA, J. D. Watson, F. H. C. Crick, M. H. F. Wilkins, and R. Franklin proposed a model for the structure of DNA in the early 1950s. They suggested that a molecule of DNA consists of two helical polynucleotide chains wound around a common axis to form a right- handed “double helix.”
In contrast to the arrangement of amino acid side chains in helical polypeptides (where the side chains are directed radially away from the helix axis), the purine and pyrimidine bases of each polynucleotide chain were directed inward toward the center of the double helix so that they faced each other.
On the basis of stereo-chemical studies, Watson and Crick further suggested that the only possible arrangement of the nitrogenous bases within the double helix that was consistent with its predicted dimensions was that in which a purine always faced a pyrimidine, for the diametric distance between the two polynucleotide chains is too small to accommodate two juxtaposed purines.
Which purine was matched with which pyrimidine became clear from a consideration of which pairs would be able to form the hydrogen bonds necessary to stabilize the double-helical structure. Accordingly, Watson and Crick concluded that adenine must be matched with thymine and guanine with cytosine.
This conclusion was, of course, in agreement with the chemical findings of Chargaff (see above)—in fact, Chargaff’s data may have been critical to the development of Watson and Crick’s proposals. The manner in which hydrogen bonds are formed between adenine and thymine and between guanine and cytosine is shown in Figure 7-6.
Although individually weak, the great number of these bonds contributes appreciably to the stability of the double-helical structure. In addition, the double helix is stabilized by hydrophobic bonds between neighboring nitrogenous bases of each polynucleotide chain. Certain other features of the structure of DNA should be noted.
The two polynucleotide chains that make up the molecule are antiparallel. That is, beginning at one end of the molecule and progressing toward the other, successive nucleotides of one chain are joined together by 3’→5′ phosphodiester linkages, whereas the complementary nucleotides of the other chain are joined by 5’→3′ phosphodiester linkages. This antiparallel arrangement is depicted diagrammatically in Figure 7-7.
A-DNA and B-DNA:
Right-handed, double-helical DNA can exist in either of two principal forms: these are called A-DNA and B-DNA. The two forms differ primarily in the positioning of the nitrogenous bases around the axis of the double helix and in the numbers of bases per helical turn. In B-DNA, there are 10 base pairs per turn of the helix, each turn sweeping out 3.4 nm (34 A) of linear translation (see Fig. 7-8); in A- DNA, there are 11 bases per helical turn, each turn sweeping out 2.8 nm (28 A) of linear translation.
Thus in the A form, the double helix has a greater diameter. In B-DNA the complementary bases lie in a plane that is perpendicular to the axis of the helix, whereas in A- DNA, the planes of successive base pairs are tilted relative to the helical axis. It is the B form of the DNA that is believed to predominate in cells, although the A form may exist in DNA-RNA hybrids.
The two polynucleotides are twisted around one another in such a way as to produce two helical grooves in the surface of the molecule; these are called the major and minor grooves (Fig. 7-8). The floor of the major groove is lined with oxygen and nitrogen atoms that could form hydrogen bonds with the amino acid side chains of proteins.
Indeed, it is the association of specific proteins with DNA that is believed to be involved in regulating gene expression. Interaction of water molecules with the atoms lining the floor of the minor groove is believed to contribute to the stability of the B form.
Replication of DNA:
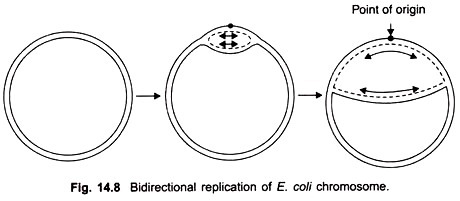
One of the intrinsic properties of the genetic material is its capacity for replication. The manner in which DNA satisfies this requirement is apparent from the nitrogenous base pairing required in the model. Because the sequence of bases in one polynucleotide chain automatically determines the sequence of bases in the other, it is clear that one-half of a molecule (i.e., one of the two helices) contains all the information necessary for constructing a whole molecule.
For example, if we know that the sequence of bases along one polynucleotide chain of DNA is A T G A C, and so on, then the complementary sequence in the other chain must be T A C T G, and so on. Therefore, if the double helix were unwound, each separate polynucleotide chain could act as a template for the production of a new, complementary chain.
The result would be two identical double helices where there was only one before. Of course, one-half of each new double helix would be represented by one of the original polynucleotide chains. The basic features of this process are shown in Figures 7-9 and 7-10. A detailed description of the mechanism by which the replication of DNA occurs, together with its experimental basis.
Denaturation and Renaturation of DNA:
DNA is readily denatured by extremes of temperature and pH. The denaturation takes the form of an unwinding of the double helix as hydrogen bonds between complementary bases are disrupted. This form of DNA denaturation is referred to as melting and produces separate DNA strands. Solutions of DNA absorb ultraviolet light (UVL) having a wavelength of 260 nm. When the temperature of a native solution of DNA is elevated, the resulting melting is accompanied by an increase in UVL absorption.
This hyperchromic effect occurs because the purines and pyrimidines of separated strands can absorb more light energy than when they are part of a double helix. Some viruses contain a single-stranded form of DNA (see later), and because this form does not exhibit the hyperchromic effect, it is readily distinguished from double- stranded forms.
When DNA is melted thermally, denaturation begins in regions of the double helix that are rich in A-T base pairs and progressively shifts to regions of greater and greater G-C content. This is because the two hydrogen bonds holding each A-T pair together can be broken more easily (hence, at a lower temperature) than the three hydrogen bonds holding each G-C pair together. A quantitative measure of the change in UVL absorbance that takes place as the temperature of a DNA solution is slowly elevated is called a melting curve (Fig. 7-11).
The point in the melting curve at which the change from double-stranded to single- stranded DNA is half complete is called the Tm value and is characteristic of a particular source of DNA. The species specificity that is characteristic of DNA melting curves reflects differences in the G-C and A-T compositions of different kinds of DNA (Fig. 7-12).
Thermally denatured DNA can be re-natured by lowering the temperature of the solution, whereby separated strands recombine to form double helices as hydrogen bonds between complementary bases are reformed. This re-annealing can be monitored as a decrease in UVL absorption by the DNA solution. The capacity for denatured DNA to re-anneal can be used to assess the size of an organism’s genome and the complexity of the DNA that is present.
When reannealing studies are to be performed, the isolated DNA is first broken by shearing force into lengths of several hundred to several thousand nucleotide pairs. The double-helical DNA is then thermally denatured yielding single strands. A known concentration of the single-stranded DNA is then incubated at the reannealing temperature (usually about 25° below the Tm) and the reannealing rate is determined from the rate of change in UVL absorbance.
A large genome reanneals more slowly than a small genome because there is a greater number and variety of DNA fragments. Thus, each fragment takes a longer time to “seek out” and anneal with its complementary partner. The kinetics of DNA renaturation (Fig. 7-12) is represented by a curve relating the percentage of reassociated fragments to the “C0t number” (i.e., the concentration of DNA in moles of nucleotides per liter (C0) times the reaction time (t) in seconds).
As seen in Figure 7-12, viral DNA (curve c) reanneals more rapidly than prokaryote DNA (curve d), and the latter reanneals more quickly than eukaryote DNA (curve e). The discovery of a eukaryotic DNA fraction in mammalian cells that reanneals unexpectedly rapidly (curve b) revealed for the first time the existence in the genomes of higher organisms of repetitive DNA sequences.
In Figure 7-12, curve shows the reannealing rate of a solution containing a mixture of synthetically produced polyuridylic acid (a nucleotide chain with only uracil bases) and poly- adenylic acid strands. Even though uracil is not usually found in DNA, like thymine it can form hydrogen bonds with adenine and does so in many RNA molecules (see below).
Z-DNA:
For about 25 years following the original establishment of the Watson – Crick Model of DNA structure, it was presumed that all naturally occurring double- helical DNA was right-handed. However, in 1979 A. Rich confirmed earlier observations reported by F. M. Pohl and T. Jovin that a left-handed form of DNA also exists. As in right-handed DNA, the two helices are held together by complementary base pairing and the strands are antiparallel. Because the sugar- phosphate backbones of the two polynucleotides trace a zigzag course around the axis of the helices (Fig. 7- 13), this left-handed DNA has been called Z-DNA.
Though the structure of Z-DNA originally proposed by Rich was based on studies of DNA crystals produced in the laboratory, Z-DNA has since been identified in the chromosomes of a number of eukaryotic cells. A variety of indirect evidence also implies that Z-DNA is a normal constituent of animal cells, plant cells, and bacteria. Unlike B-DNA,’Z-DNA is highly immunogenic, making it possible to readily produce antibodies against Z-DNA. The reaction of these antibodies with DNA isolated from a variety of cell types implies the presence of the Z form. Naturally occurring Z-DNA-binding proteins have also been isolated from a number of cells.
The Z form of DNA appears to coexist with the B form in the same DNA molecules. Indeed; it is believed that DNA can “flip” between the B and Z forms in those regions of a double helix that are rich in sequences having alternating purines and pyrimidines. These sequences appear to occur in selective regions of a cell’s DNA, lending credence to the idea that switches in DNA helicity between right-handed and left-handed forms may be involved in selective gene expression.
“Single-Stranded” DNA:
Although in nearly every case so far studied, DNA consists of two polynucleotide chains twisted about one another to form a double helix, it is now apparent that in a few bacterial viruses (i.e., the φ X 174 and S13 E. coli phages) DNA exists as a single polynucleotide chain. This was initially suspected when chemical analyses of the nitrogenous base contents of these viral DNAs revealed that the amounts of adenine and thymine, as well as guanine and cytosine, were not equal.
During reproduction of these viruses, the single-stranded DNA (referred to as the ” + strand”) is injected into the host bacterial cell, where it acts as a template for the reproduction of a complementary polynucleotide chain (called the “- strand”); these two polynucleotides combine to form a conventional double helix, which then serves as the template for the production of additional + strands. The newly produced + strands are then enclosed in the viral protein coats to form new virus particles.
Structure of RNA:
RNA and DNA differ chemically in two notable ways: in RNA, ribose is the pentose (not deoxyribose as in DNA) and the pyrimidine uracil occurs in place of thymine (Fig. 7-14). Early chemical analyses of the nitrogenous base contents of RNAs from various sources revealed that the A:U and G:C molar ratios were quite different from the A:T and G:C molar ratios of DNA. On this basis, it was concluded that RNA occurs as a single polynucleotide chain.
This contention is also supported by physico- chemical studies, but it should be noted that there are some viral RNAs that are double stranded. Although only one polynucleotide chain is usually present, RNA does possess regions of double-helical coiling where the single chain loops back upon itself.
These regions are stabilized by the formation of hydrophobic bonds between neighboring bases (as in DNA) and also by the formation of hydrogen bonds between guanine and cytosine and between adenine and uracil.
In RNA, A and U can form two hydrogen bonds similar to the two bonds formed between A and T in DNA. As in DNA, the portions of the RNA strand that are twisted around each other to form a double helix are antiparallel. In double-helical RNA, the helices and the complementary base pairs are arranged in much the same manner as in A-DNA.
Synthesis of RNA:
Except perhaps in the case of the reproduction of certain RNA viruses, the synthesis of RNA appears ‘to be directed by DNA and is called transcription. The formation of the RNA polynucleotide takes place using the base sequence along only one of the two deoxyribonucleotide helices of DNA (producing a temporary RNA-DNA hybrid) as a template and results in the release of a single, complementary polyribonucleotide chain in which the base uracil occurs in place of thymine (Figure 7-14).
Replication of DNA and RNA Viruses:
The viruses may be divided into two classes: viruses whose genetic complement consists of DNA and viruses whose genetic complement consists of RNA. In cells infected with DNA viruses, the infecting viral DNA is replicated, forming new viral DNA that is then transcribed into RNA; this RNA is then translated into viral protein (Fig. 7-15a).
The newly produced viral DNA and viral proteins combine in the assembly of new, complete virus particles that are released upon lysis of the host cell. A latent state can also be established in which the viral DNA is incorporated into the host cell’s genome, being replicated and distributed along with the host cell’s native DNA, until it is transcribed once again into additional viral RNA and then into viral proteins.
For most RNA viruses (e.g., poliomyelitis, influenza, common cold, etc.), DNA involvement is essentially bypassed. For example, during the infection of a cell with polio virus, the single-stranded RNA (called a + strand) enters the host cell, where it acts as a template for the synthesis of complementary – strands. The latter are then employed in the proliferation of new + strands and these are translated into viral proteins (Fig. 7-15b).
The mechanism described above is varied in several other viruses in which the RNA is either double stranded (e.g., reoviruses, in which only one of the two RNA strands produced during replication is transcribed) or the infecting single RNA strand is complementary (rather than identical) to the newly produced viral RNAs that are to be translated into viral proteins (e.g., Sendai virus, Newcastle disease virus, etc.)
Yet another mechanism exists in the case of the RNA tumor viruses (e.g., Rous sarcoma virus). These viruses do not transfer information from RNA to RNA, but rather from RNA to DNA and then to new RNA. The viral RNA is employed as a template for the synthesis of DNA by the infected cell (a phenomenon that is called “reverse transcription”). Some of the resulting “viral DNA” may be incorporated into the genome of the host cell, establishing what is called the provirus state.
The provirus state has been suggested as the basis of a number of different RNA virus-induced and DNA virus-induced cancers. According to this view, one or more of the provirus genes—which are normally repressed by the host cell—may become derepressed and cause the production of an oncogenic (i.e., cancer- causing) substance that alters the cell’s normal properties or behavior. Such a change may be delayed for a number of generations, depending on the period of latency.
During the 1960s and early 1970s, the so-called “central dogma” of molecular biology was the orderly and unidirectional flow of information encoded in the base sequences of a cell’s DNA to RNA and then to protein, that is,
The discovery of reverse transcription by certain RNA viruses in which the information of RNA is passed on to DNA has necessitated a reexamination of that dogma and raised the question of whether a similar interaction between RNA and DNA might normally occur in cells (i.e., cells not infected by viruses) under specific conditions (e.g., during cellular differentiation). The central dogma might more appropriately be represented as,
Types of Cellular RNA:
Cells contain three major functional types of RNA: ribosomal RNA (abbreviated rRNA), messenger RNA (mRNA), and transfer RNA (tRNA). All these are transcribed from DNA and are engaged in mediating the expression of the genetic message of DNA by participating in the synthesis of the cell’s proteins. It has already been noted that RNA occasionally serves as the genetic material of viruses.
Of the cellular RNAs, rRNA is the most abundant, accounting for up to 85% of the total RNA of the cell. Only three “or four- different kinds of rRNA are present in cells, and these are confined for the most part to the cell’s ribosomes. mRNA accounts for about 5 to 10% of the cell’s RNA and is much more heterogeneous with respect to size and nitrogenous base content than the rRNAs.
This results from the relationship (see below) between the chain lengths and base sequences of mRNAs and the variable sizes and primary structures of polypeptides synthesized in a cell. Most mRNA occurs in the cytoplasm, where it transiently combines with ribosomes during protein synthesis.
About 10 to 20% of the cell’s RNA is tRNA. All tRNA molecules are similar in size and typically contain 75 nucleotide units. In spite of these similarities, a single cell may contain about 60 species of tRNA differing in their base sequences. Because most of the tRNA is recovered in the cytoplasmic (i.e., soluble) phase of disrupted cells following centrifugation, tRNA is also called soluble RNA (i.e., sRNA).
In view of its small size and relative ease of isolation, tRNA has been more extensively studied than the other two ribonucleic acids, and the specific primary, secondary, and tertiary structures of many tRNAs have already been determined. tRNAs contain moderate amounts of unusual nucleotides such as ribothymidine, dihydrouridine, pseudouridine, and methylguanosine. These are formed by modification of the four common RNA bases after the tRNA has been synthesized from the four unmodified ribonucleotides. The modified bases play a crucial role in establishing the unique spatial organization of these molecules.
Although the mechanisms of DNA replication and protein synthesis are considered in depth, it is appropriate that a brief accounting of the functional relationships among the nucleic acids and between nucleic acids and proteins be made at this time. Inheritable information is encoded in the various nitrogenous base sequences possessed by the cell’s DNA, and by the process of transcription and translation these base sequences are employed to specify the primary structures of all proteins, produced by the cell.
Most important among these proteins are the enzymes that catalyze and regulate the myriad of chemical reactions characterizing the cell’s metabolism. Therefore, the information of DNA confined essentially to the cell nucleus manifests itself primarily in the cytoplasm as the synthesis of a unique assemblage of proteins.
The replication of DNA that precedes mitotic cell division and the equal distribution of the duplicated DNA among the progeny cells provides for the passage of complete sets of information from one generation of cells to another. In addition to serving as templates for their own replication, the nucleotide sequences of DNA are used during transcription to produce complementary base sequences of RNA.
The resulting RNAs then serve as intermediaries in translating the original message into protein. Of paramount importance in this process are the mRNA molecules whose base sequences directly determine the primary structures of the polypeptides. These mRNA molecules leave the nucleus of the cell following their synthesis and attach in the cytoplasm to one (or several) ribosome.
The rRNA of each ribosome is believed to play a role in this attachment. tRNA molecules also produced in the nucleus enter the cytoplasm, where they combine with specially activated amino acids (distinct tRNAs exist for each species of amino acid). The resulting complexes, directed by the base sequence of mRNA attached to the ribosomes, sequentially deposit their amino acids in the growing polypeptide chains.