The following points highlight the three main stages for translation of RNA and protein synthesis. The stages are: 1. Initiation of Polypeptide 2. Elongation of Polypeptide: 3. Termination of Polypeptide.
Translation of RNA: Stage # 1. Initiation of Polypeptide:
Ribosomes exist as separate large and small subunits. The first step in translation involves the binding of the small ribosomal subunit to the mRNA. Translation usually begins at the sequence AUG, sometimes GUG, which encodes methionine and is known as the translation initiation codon.
The small subunit binds to the mRNA at a specific point (Shine-Dalgarno sequence) upstream of the AUG. In eukaryotes, the small ribosomal subunit recognizes the cap structure at the 5′ end of the mRNA. It then moves downstream until it encounters the first AUG. A tRNA charged with methionine binds to the AUG located by the small ribosomal sub-unit.
In Prokaryotes:
Initiation of polypeptide chain occurs with the involvement of initiation factors, ribosomal subunits and amino acyl-tRNA complex.
Initiation factors:
A number of accessory proteins called initiation factors are required for initiation. Bacteria have three, known as IF1, IF2, and IF3. Initiation begins with binding of IF1 and IF3 to the small ribosomal subunit. This helps to prevent binding of a large subunit before the mRNA has bound. Next, IF2 complexed with GTP binds the small subunit. Its purpose is to assist binding of the initiator tRNA.
The small subunit then binds the mRNA and locates the AUG initiation codon. The initiator tRNA charged with methionine binds to the complex and IF3 is released. Then large ribosomal subunit binds to the initiation complex to from a complete functional ribosome. This is accompanied by the release of IF1 and IF2 and hydrolysis of GTP (Fig. 16.10).
Formation of Formyl-methionyl tRNAƒmet:
In prokaryotes,. methionine carries a formyl group (-CHO) and hence it is called N-formyl methionine. The’ initiation of synthesis takes place through initiation tRNA. It is abbreviated as tRNAƒmet. The initiation tRNA or tRNAƒmet forms a complex with methionine called methionyl tRNAƒmet.
The amino group of methionine is blocked by formyl group to form N formyl methionyl tRNAƒmet. This reaction is catalysed by a transformylase.
Binding of Initiation factors with 305:
Small ribosomal subunit binds IF1, IF3. Next, IF2 complexed with GTP binds the small subunit.
Binding of 30S with mRNA:
Ribosomes are the sites of protein synthesis and are found dissociated into subunits (505 & 305). The 305 subunit attached at the AUG codon of the mRNA forming the mRNA-30S complex. The process requires the initiation factor IF3.
Binding of the fmet-tRNAfmet with 305- mRNA Complex:
The fmet-tRNAfmet attaches with the 30S-mRNA complex to give rise to the initiation complex 305-mRNA-fmet-tRNA/met. This reaction is facilitated with initiation factor IF2. IF3 is released in this step.
Association of Ribosomal Subunits:
The initiation complex formed then associates with SOS subunit to reconstitute 70S ribosome. During this process OTP is converted to GDP and the initiation factor IF1 and IF2 are released.
In Eukaryotes:
There are minor differences in the initiation of polypeptide chains In between prokaryotes and eukaryotes.
Initiation Factors:
In eukaryotes there are more Initiation factors. These factors are elF1, elF2, elF3, elF4A, elF4B, elF4C, elF4D, elF4F, elF5, elF6. The elF2, elF3, elF4A and elF4F contain multiple polypeptide chains, but others are simple polypeptides; elF2 and elF3 are analogous to IF2 and IF3 of prokaryotes.
Formation of Ternary Complex:
OTP binds to elF2 which increases its affinity for met- tRNAmet. This met-tRNAmet associates with elF2 – GTP complex forming a ternary complex, i.e., met-tRNAmet-elF2-GTP. The initiator tRNA is not formylated in eukaryotes.
Association of Ternary Complex with 40S Subunit:
The ternary complex associates with 40S subunit, to form 43S initiation complex. The factor elF2 has three subunits, namely α, β, γ. The elF2α binds to GTP, elF2γ binds to met-tRNA and elF2βmay be a recycling factor.
Binding of mRNA with 43S Initiation Complex:
The mRNA at its 5′ end binds with 43S initiation complex. This reaction depends on elF3 and the binding of mRNA is assisted by elF4F, elF4B and a high energy bond of ATP.
Association with 60S Subunit:
After association of the S’ end of mRNA, the initiation complex moves towards 3′ end in search of initiation codon AUG and then also associates with 60S subunit.
This association of 60S subunit with the initiation complex requires the factor elFS, because it helps in releasing elF2 and elF3. elF2 is released as a binary complex, elF2-GDR The 40S-60S joining reaction really depends on elF4C, and the GTP of the initiation complex is hydrolysed, when SOS ribosome is reconstituted.
Translation of RNA: Stage # 2. Elongation of Polypeptide:
Elongation of polypeptide has been represented in Fig. 16.11.
Elongation Factors:
A number of accessory proteins are required for elongation. In prokaryotes, two elongation factors, EF-Tu and Ef-Ts, are involved. EF-Tu is associated with entry of a tRNA into the A site of ribosome. Ef-Tu binds charged tRNAs in association with GTP. Following entry to the A site, the GTP is hydrolyzed and EF-Tu is released bound to guanosine diphosphate (GDP).
Before another tRNA can bind, EF-Tu must be regenerated with the help of Ef-Ts.
First, EF-Ts displaces GDP by binding to EF-Tu; a new molecule of GTP then replaces EF-Ts (Fig. 16.12). In eukaryotes, a complex protein called eEF-1 brings the tRNA to the A site. Again, the reaction is associated with hydrolysis of GTP.
Binding of AA-tRNA at A Site of Ribosome:
The larger subunit of ribosome has two sites where two molecules of tRNA are attached. These are called P site (peptide site) and A site (aminoacyl site). fmet-tRNAƒmet at first comes to the A site, then to the P site to make the A site available for the next aminoacyl tRNA. For the attachment of AA-tRNA to A site, a molecule of GTP and elongation factors Ef-Tu and EF-Ts are required.
Ef-Tu forms a ternary complex with AA-tRNA and GTP (AA-tRNA-GTP- EF-Tu). The formation of this complex is catalysed by EF-Ts. These complex deposits AA-tRNA at A site and GDP, EF-Tu are released along with the phosphate group.
Formation of Peptide Bond:
This is a catalytic reaction during which a peptide bond is formed between the free carboxyl group (-COOH) of the peptidyl tRNA at the P site and the free amino (-NH2) group present with amino acyl tRNA at the A site.
The enzyme involved in this reaction is peptidyl transferase and is an internal part of SOS ribosomal subunit. After the formation of peptide bond, the tRNA at P site is de-acylated and the tRNA at A site carries the polypeptides.
Translocation of Peptidyl tRNA from A to P Site:
The peptidyl tRNA present at A site is then trans-located to P site.
For translocation of peptidyl tRNA from A site to P site, there are two models:
(i) According to two sites (A, P) model, de-acylated tRNA is liberated from P site, and with the help of one GTP molecule and an elongation factor EF-G, the peptidyl tRNA is trans-located from A to P site. The elongation factor EF-G, binds to ribosome, is released on hydrolysis of GTP.
(ii) According to three sites (A, P, E) model, initially the aminoacyl end of the t-RNA bound to the A site moves to the P site on SOS subunit at the time of peptide transfer, but only later during translocation, the anticodon end of this tRNA moves from A to P site on 30S subunit. This latter step requires action of elongation factor EF-G.
In this model, thus, there is an intermediate state, when anticodon end of this tRNA is still on A site (on SOS sub- unit), while the aminoacyl end occupies P site (on SOS subunit). A third tRNA binding site, E was also recognized on SOS subunit, through which tRNA leaves the ribosomes.
Translation of RNA: Stage # 3. Termination of Polypeptide:
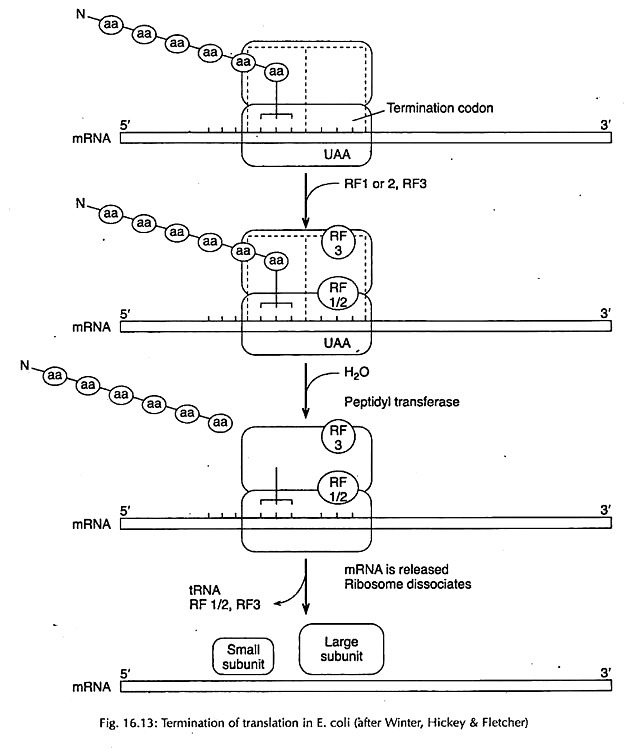
Termination of the polypeptide chain is brought about by the presence of any one of the three termination codons, namely UAA, UAG and UGA.
Release Factors:
In prokaryotes, termination codons are recognized by one of the two release factors, RFl and RF2. Of these release factors, RF1 recognizes UAA and UAG and RF2 recognizes UGA. They help the ribosome to recognize these triplets. A third release factor RF3 seems to stimulate the action of RF1 and RF2. In eukaryotes, there is only one release factor (eRFI).
Release of Polypeptide:
For release action, the poly-peptidyl tRNA must be present on P site and release factors help in splitting of the carboxyl groups between the polypeptide and the last tRNA carrying this chain (Fig. 16.13). Polypeptide is thus released and the ribosome dissociates into 2 subunits with the help of IF3.
In eukaryotes, there is only one release factor – eRFI. GTP seems to be necessary for binding of this factor to ribosome. GTP is deducted after the termination step has occurred which may be a prerequisite for the release of RF1 from the ribosome.
Modification of the Released Polypeptide:
The released polypeptides are modified in various ways. An enzyme deformylase removes the formyl group of first amino acid methionine. Due to the action of certain other enzymes, exo-amino-peptidases, amino acids may be removed a tertiary structure. In this manner, these proteins from either the N-terminal end or the C-terminal ultimately become functional enzymes, end or both.
The polypeptide chain singly or in association with other chain also folds to take up a tertiary structure. In this manner, these protein ultimately become functional enzymes. The whole process of translation is present association with other chain also folds to take up ted in Fig. 16.14.