The following points highlight the three main classes of RNA molecules and RNA processing. The classes are: 1. Messenger RNA 2. Ribosomal-RNA 3. Transfer-RNA.
Class # 1. Messenger RNA:
Messenger RNA (m-RNA) is an informational molecule. The genetic message for protein synthesis contained in DNA is transcribed in the complementary ribonucleotide sequence of m-RNA. A group of three consecutive nucleotides specifies one amino acid in the polypeptide chain of a protein.
Naturally, the size of a messenger molecule depends on the size of the polypeptide coded by it. An average-sized polypeptide containing about 300-600 amino acid residues is coded by a m-RNA consisting of 900-1800 nucleotides.
m-RNA molecules are generally short-lived. Prokaryotic messengers have an average half-life of about 2 minutes, while the eukaryotic ones of about 10 minutes. The significance of such short life of m-RNAs is that when the synthesis of a particular protein is not needed, it can be stopped by turning off transcription of the particular gene. The already synthesized m-RNA is rapidly degraded by RNase, so that synthesis of a protein that is not needed may be stopped within a short time.
Eukaryotic messengers are transcribed in the form of a much longer precursor, the hn-RNA, containing extra segments which are excised to produce the actual m- RNA– that is translated.
In contrast, the prokaryotic messengers do not have such extra segments or introns. Eukaryotic messenger in the form of hn-RNA may have few to many (as many as 56) introns in one molecule. The size of the introns is also highly variable — ranging from 80 nucleotides to 10,000, or even more. Each intron contains conserved sequences at 5′- and 3′-splice sites, i.e. the sites at which the intron is excised from hn-RNA.
This excision of introns and joining of exons to produce a functional RNA molecule are known as RNA-splicing. The splicing reaction requires ATP and several small ribonucleoproteins (sn-RNP) present in the cell nucleus. The hn-RNA, together with the sn- RNP molecules, assemble into a large ribonucleoprotein complex, known as a spliceosome. Splicing of an hn-RNA is diagrammatically shown in Fig. 9.32.
The spliceosomes seem to function by recognizing the consensus sequences at the intron splicing sites. The splicing mechanism ensures that each 5′-splicing site pairs with the nearest downstream 3′-splicing site, so that the intervening sequences of only one intron is excised at a time.
This mechanism prevents removal of any exon lying in between the introns as shown in a simplified manner in Fig. 9.33:
Class # 2. Ribosomal-RNA:
In both prokaryotes and eukaryotes, a number of different ribosomal RNAs are present as components of ribosomes. Ribosomes are large RNA-protein complexes which are visible under an electron microscope. They perform an important function in protein synthesis. In prokaryotic cells they are present in the cytoplasm. In eukaryotes, they are present both in the cytoplasm and on the surface of the endoplasmic reticulosystem, as well as in mitochondria and chloroplasts.
The prokaryotic ribosomes are somewhat smaller than the eukaryotic ones. They have a sedimentation coefficient of 70S and consist of two subunits, 50S and 30S. The 70S particles have a molecular weight of 2500,000 Daltons and the 50S and 30S subunits have molecular weights of 1,600,000 Daltons, and 900,000 Daltons respectively. The 50S subunit contains two types of RNA molecules. 5S and 23S having 120 and 2,900 nucleotides, respectively, and 34 different protein molecules.
Each 50S subunit contains one copy of each of two types of RNA molecules. The 30S subunit contains only one type of RNA (16S) having 1,540 nucleotides, and 21 different protein molecules. It is interesting to note that the mitochondrial and chloroplast ribosomes of eukaryotes are also 70S particles which suggests their prokaryotic origin.
The cytoplasmic and endoplasmic reticulum-bound ribosomes of eukaryotes are 80S particles composed of a 60S and a 40S subunits. The 60S subunit (M.W. 2,800,000 Daltons) has three types of RNA molecules, — 5S (120 nucleotides), 5.8S (160 nucleotides) and 28S (4,700 nucleotides) and 48 different protein molecules. The smaller 40S subunit (M.W. 1,400,000 Daltons) is composed of a single kind of RNA (18S) of 1,900 nucleotides and has a molecular weight of 1,400,000 Daltons. The 40S subunit also contains 33 different protein molecules.
The characteristics of prokaryotic and eukaryotic ribosomes are summarized in Table 9.2 :
An important feature of the ribosomal RNAs of both prokaryotes and the eukaryotes — except the 5S RNA of eukaryotes — is that they are transcribed together as a composite transcript. In E. coli the three species of r-RNA: 5S, 16S and 23S RNA, are synthesized together in the form of a primary transcript which contains, besides these, also some transfer RNAs.
One such composite primary transcript is shown:
The composite primary transcript needs processing to separate the three r-RNAs and the four t-RNAs. As the three types of RNAs are produced from the same transcript, their ratio remains 1:1: 1. This is significant, because each prokaryotic ribosome contains only one molecule of each of the three types.
In case of eukaryotes, the 5S RNA is transcribed separately, while the three others, 5.8S, 18S and 28S r-RNAs are produced from a composite transcript. The eukaryotic 5S r-RNA is transcribed from a cistron containing 120 bp. Several such cistrons are present in a row separated by non-transcribed sequences, known as spacers. Other eukaryotic r-RNA cistrons encoding 5.8S, 18S and 28S r-RNA’s are also provided with spacers. The length of the spacer sequences is variable. In case of 5S r-RNA the spacer contains 600 bp.
Another characteristic feature of eukaryotic r-RNA cistrons is that they are present in multiple copies. For example, in human cells, there are about 100 copies of these genes in a haploid set of chromosomes. Such multiple copies of r-RNA genes are also present in bacteria, though the number of copies is much less. For example, in E. coli there are only seven copies. The number is very high in case of amphibians. For example, in Xenopus, the number is as many as 600.
In eukaryotes, r-RNAs soon after their processing are utilized to build the ribosomes, a process known as packaging. This packaging is done in the nucleolus. The nucleolus contains large loops of DNA protruding from some chromosomes and containing the multiple copies of r-RNA genes. These chromosomes are known as nucleolus organizers.
In the ciliated protozoan, Tetrahymena, the processing of r-RNA takes place by a unique splicing mechanism carried out by a non-proteinaceous catalyst known as ribozyme. The r-RNA molecules of this organism are transcribed initially as a large precursor molecule containing exons and introns from which one RNA molecule is produced by RNA-splicing reaction.
Surprisingly, it was discovered that such splicing can be carried out in vitro in absence of any protein enzyme. The intron sequence itself can catalyse the splicing reaction. The 400 base-pair long intron sequence can function as an enzyme. Since, the intron is also RNA the catalyst is known as ribozyme. It was the first evidence to show that RNAs could also have enzymatic activity. Since then, ribozymes having self-splicing activity have been discovered in other systems, including fungi and bacteria.
The mechanism of self-splicing is explained schematically in Fig. 9.34:
Class # 3. Transfer-RNA:
Transfer-RNAs (t-RNA) are the smallest among the three classes of RNAs. They remain soluble in the cytosol and are also called soluble-RNA. They function as adapter molecules, because, on one hand, they carry an amino acid and, on the other, have a specific sequence of three nucleotides (anticodon) by which they recognize a specific codon on the ribosome-bound m-RNA.
Thus, each t-RNA carries a specific amino acid to a particular site on m-RNA at which a peptide bond binds the incoming amino acid to adjacent one during protein synthesis. As most proteins contain variable number of 20 different amino acids (protein amino acids), and each amino acid is transferred by a specific t-RNA, there must be at least 20 different types of t-RNA molecules.
In fact, each cell has about 50 different t-RNAs to carry the 20 different amino acids. This is necessary because most of the amino acids are coded by more than one codon (degeneracy of the genetic code). To match the m-RNA codons, anticodons of equal number of different t-RNAs are necessary.
Although, like ribosomal or messenger RNAs, t-RNAs are single-stranded, they possess a characteristic feature in having a secondary structure due to formation of double-stranded portions and loops by intrastrand base-pairing. As a result they possess a three-dimensional configuration, commonly known as the clover-leaf structure.
Such a structure is common to all t-RNAs, although the base sequences are different. The clover-leaf structure shown in Fig. 9.35 is produced by intrastrand base- pairing and loops of unpaired bases (leaflets). All t-RNAs also have a common terminal sequence consisting of three nucleotides -C-C-A at the 3′-(OH) end.
The 5′-end contains a G (guanylic acid residue). Another notable feature of all t-RNAs is the presence of several unusual bases, like methylated derivatives of A, U, G, and C, pseudo-uracil and di-hydro-xyuracil. Each t-RNA also contains an anticodon in the central leaflet and the anticodon triplet is flanked on either side by a U and a methyl pyrimidine.
The 3′-(OH) C-C-A is the amino acid binding site. The carboxyl group of an amino acid is linked with either the 2′ or 3′ (OH) group of the last nucleotide i.e. adenylic acid by an ester bond. The base- triplet at the anticodon site determines the amino acid to be carried by the t-RNA. The anticodon triplet forms H-bonds with the codon triplet of the m-RNA, so that the amino acid carried by the specific t-RNA can be correctly positioned for transfer to the growing polypeptide chain.
Thus, each t-RNA has a specific sequence in the anticodon site. From a pool of t-RNAs and amino acids, a specific t-RNA picks up its correct amino acid and with the help of its unique anticodon, brings the amino acid to its proper place on the ribosome-bound m-RNA, where the anticodon matches with the specific codon of the amino acid.
Though t-RNA molecules are depicted as flat structures as shown in Fig. 9.35 for the sake of simplicity, they have a more complicated three-dimensional folded structure as revealed by X-ray crystallography. The folded molecule assumes an L-shape. The amino acid binding C-C-A-end stands at one end of the L-shaped molecule and the anticodon loop at the other end.
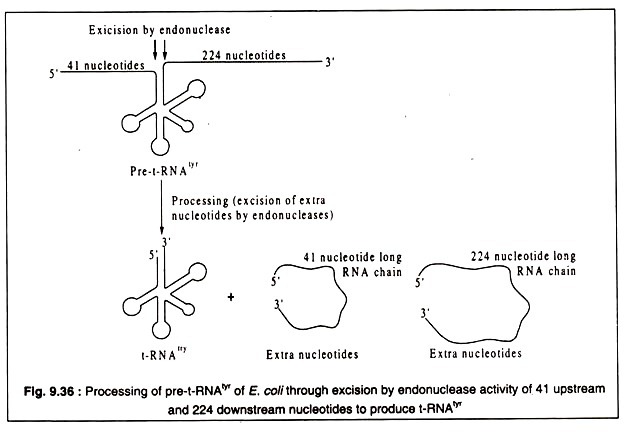
All t-RNAs of both prokaryotes and eukaryotes are processed from much longer primary transcripts which are called pre-t-RNAs. For example, the E. coli t-RNA specific for tyrosine (t-RNAtyr) which is 85 nucleotide long after processing is produced from a pre-t-RNAtyr which has 350 nucleotides. It has 41 extra nucleotides at the 5′-end and 224 extra nucleotides at the 3′-end. These extra nucleotides are removed by endonucleases to produce the correct t-RNAtyr of E. coli. This is diagrammatically shown in Fig. 9.36.
The endonuclease which removes the 41 base sequences at the 5′-end of pre-t-RNAtyr of E. coli is unusual in having 86% RNA and 14% protein. It is called RNAse-P. The protein part helps only to maintain a correct conformation. The catalytic activity of the endonuclease resides in the RNA part. Thus, RNAse-P is a ribozyme. Similar ribozymes have been found to be involved in the processing of eukaryotic t-RNAs.
All t-RNAs contain some unusual bases. These are produced by modification of the usual nucleic acid bases. The modification is accomplished by specific enzymes after the t-RNAs are processed to their final form.
In yeast, a simple eukaryotic organism, the primary transcript of t-RNA genes may contain segments transcribed from introns which vary in length from 10 to 30 nucleotide sequences. These portions of the pre-t-RNAs are removed and the exons are joined to yield mature t-RNAs. Another difference from prokaryotic t-RNAs is that the -C-C-A sequence at the 3′-OH group is not present initially, but is added later to obtain the mature t-RNA.
The pre-t-RNAtyr and the t-RNAtyr of yeast are shown in Fig. 9.37: