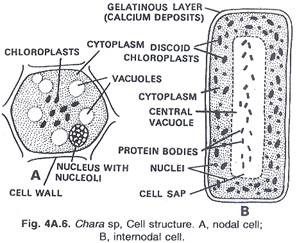
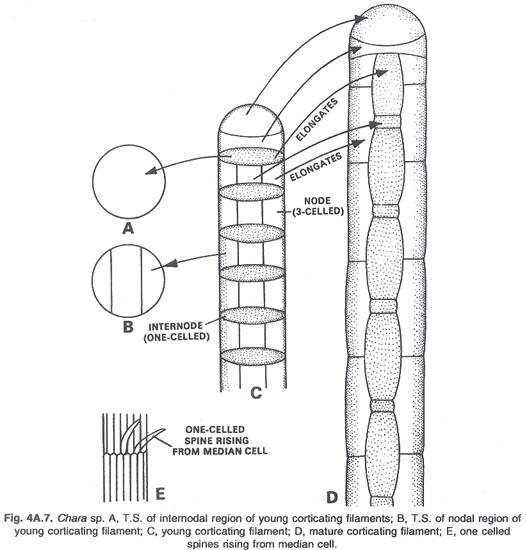
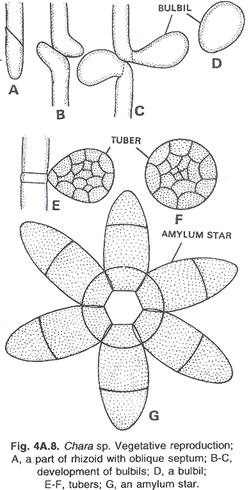
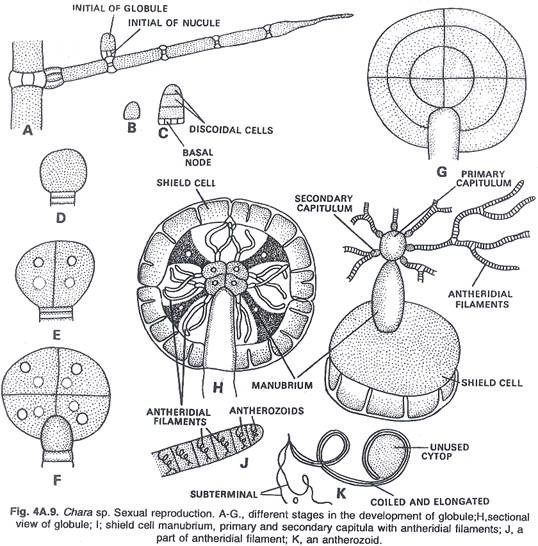
In this article we will discuss about the role of RNA polymerase in transcription.
RNA polymerase enzymes are complex enzyme which in E. coli is made up of 5 subunits or polypeptide chains designated β, β’, α, σ and ω with respective molecular weights of 160,000, 150,000, 90,000, 40,000 and 10,000. The α chain is present twice, others only once. The active form of the enzyme is called holoenzyme and has a total molecular weight of 500,000.
The polypeptide chains are held together by secondary non-covalent bonds. Chains β, β’, α and ω form the core enzyme; σ (sigma factor) is weakly attached to the other chains and can easily become detached. The core enzyme catalyses the linkage of ribose nucleotides by phosphodiester bonds.
The sigma factor recognises start sequences in the promoter region of DNA where transcription begins. In the presence of the sigma factor the holoenzyme binds to those nucleotides in promoter region which initiate transcription. Soon thereafter, the double helix unwinds and one strand serves as the template for transcription.
The eukaryotic RNA polymerases I, II and III consist of 8 to 14 different subunits in each. They recognise different promoters and recognise different classes of genes.
The two largest subunits of all three eukaryotic RNA polymerases are related to the β and β’ subunits of the E. coli polymerase. Five subunits of the eukaryotic RNA polymerases are common to all the three enzymes. These structural similarities allow eukaryotic polymerases to share several common functional properties.
Site of Transcription, Promoter Recognition:
The DNA sequence to which RNA polymerase binds to initiate transcription of a gene is called the promoter. The promoter is a specific site at the beginning of genes where transcription is initiated.
The initiation process is important because this is the primary step at which transcription is regulated. Typical promoters are from 20 to 200 bases long. The base sequence among different promoters shows variation, and that relates with the strength in binding with the RNA polymerase.
Transcription raised the question as to whether one or both strands of DNA duplex are transcribed. In virus φ X 174 which has only single-stranded DNA (unlike most other DNA viruses), only one strand of DNA is transcribed into a complementary RNA sequence. In other viruses with duplex DNA, when we consider the entire genome, both strands of DNA duplex are transcribed, one strand serving as template for some genes, the other strand for remaining genes.
Transcription in Prokaryotes:
Promoters in bacteria are located in the region of a DNA strand just preceding the initiation site of RNA transcription. The nucleotide at which transcription is initiated is denoted as + 1 and the preceding nucleotide as -1. The portions of the DNA preceding the initiation site, toward the 3′ end of the template are said to be upstream of that site.
Those portions of the DNA succeeding it, toward the 5′ end of the template are said to be downstream of that site. Comparisons of promoter sequences of a series of different genes isolated from E. coli revealed that the region upstream of the transcription initiation site contains two sets of sequences that are similar in a variety of genes.
These two common sequences, referred to as consensus sequences, contain 6 nucleotides each. Each base in the consensus sequence is the base most often observed at that position among any number of observed sequences.
Consensus sequences are located approximately 10 and 35 base pairs upstream of the transcription start site. They are called the -10 and -35 elements, denoting their position in relation to the transcription initiation site, which is at + 1 position. (Fig. 15.6).
The consensus sequence in position 35 in E. coli has TTGACA and that in position 10 has TATAAT. The sequences at the – 10 and – 35 positions are not identical in different promoters, but similar enough to establish consensus sequences. The – 10 sequence which is called the TATA box or Pribnow box (after the name of its discoverer) is similar to sequences found at corresponding positions in many eukaryotic promoters.
The positions of the promoter sequences determine where and on which strand the RNA polymerase begins synthesis. Experimental evidence supports the functional importance of the – 10 and – 35 promoter elements.
First, genes with promoter elements that differ from the consensus sequences are transcribed less efficiently than genes whose promoters match the consensus sequence more closely.
Second, mutations induced in either the – 35 or – 10 consensus sequences have strong effects on promoter function.
Third, the sites at which RNA polymerase binds to promoters have been directly identified by foot-printing experiments, widely used to determine the sites at which proteins bind to DNA (Fig. 15.7).
In these experiments, a DNA fragment is radiolabeled at one end. The labelled DNA is incubated with the protein, for example RNA polymerase, and then subjected to partial digestion with DNase. The method is based on the principle that the regions of DNA to which the protein binds are protected from DNase digestion. A parallel sample of DNA that was not incubated with protein is digested with DNase.
The regions of DNA with bound protein can be identified by comparison of the digestion products from protein-bound DNA and DNA without protein. Such foot-printing analysis has shown that RNA polymerase generally binds to promoters over approximately a 60 base pair region, extending from – 40 to + 20, that is, from 40 nucleotides upstream to 20 nucleotides downstream of the transcription start site. The sigma (σ factor) subunit of RNA polymerase binds specifically to sequences in both the – 35 and – 10 promoter regions, indicating the importance of these regions in promoter function.
Initiation of Transcription:
In absence of sigma subunit, RNA polymerase can bind non-specifically to DNA with low affinity. Sigma plays a crucial role in directing the polymerase to promoters by binding specifically to both the – 35 and – 10 sequences, leading to initiation of transcription at the beginning of a gene. The initial binding between the polymerase and promoter is referred to as closed-promoter complex because the DNA is not unwound.
The polymerase then unwinds about 15 bases of DNA around the initiation site to form an open-promoter complex and single stranded DNA is made available for transcription. The first nucleoside triphosphate is placed at the site. The next nucleotide in line is joined to the 3′ carbon of the ribose, and so forth. Transcription is initiated by the joining of two free NTPs at the + 1 site.
Chain Elongation:
After addition of about the first ten nucleotides, sigma is released from the polymerase shown in Fig. 15.6. The polymerase then moves along the DNA template strand, adding nucleotides to the 3′ end of the growing RNA chain. Thus RNA chains grow in the 5′ to 3′ direction similar to what is observed in DNA synthesis.
As it moves, the polymerase unwinds the template DNA over about 17 base pairs (less than two turns of the double helix) in the region of transcription. After RNA polymerase has passed, the DNA strands reform the duplex.
Chain Termination:
The RNA chain continues to grow until the RNA polymerase encounters a termination signal. Then transcription stops, the RNA chain is released from the polymerase, and the enzyme dissociates from its DNA template. The most common type of termination signal in E. coli consists of a symmetrical inverted repeat of a GC-rich sequence followed by 4 or more A residues.
Transcription of the GC-rich inverted repeat produces a segment in the growing RNA chain that can form a stem loop structure by complementary base pairing (Fig. 15.8).
The formation of a self-complementary structure dissociates the chain from the DNA template and terminates transcription. There are other types of transcription termination signals in prokaryotes as well as eukaryotes that involve binding of proteins to specific DNA sequences for chain termination, instead of formation of the stem-loop structure in RNA.
Transcription in Eukaryotic:
There are two major differences between prokaryotic and eukaryotic transcription systems.
First, a single RNA polymerase is able to transcribe all genes in bacteria, whereas eukaryotic cells have multiple different RNA polymerases that transcribe distinct classes of genes.
Second, eukaryotic RNA polymerases do not bind directly to promoter sequences, but interact with a number of proteins to specifically initiate transcription. The complexity of the transcription process in eukaryotes is presumed to be related with the regulation of gene expression required to control activities of many different cell types in multicellular forms.
Three distinct nuclear RNA polymerases transcribe different classes of genes in eukaryotic cells (Table).
RNA polymerase II transcribes protein coding genes in nucleus to yield mRNAs; RNA polymerases I and III transcribe ribosomal RNAs (rRNAs) and transfer RNAs (tRNAs). The three largest species of tRNAs are transcribed by RNA polymerase I.
The genes for transfer RNA and the smallest species of ribosomal RNA (5S rRNA), as well as some small nuclear (snRNAs) and cytoplasmic RNAs (scRNAs) involved in splicing and protein transport are transcribed by RNA polymerase III. In addition, mitochondria and chloroplasts contain separate RNA polymerases similar to bacterial RNA polymerases that specifically transcribe DNA in these organelles.
Transcription by RNA Polymerase II:
The different mode of action of transcription in eukaryotic cells was noted in 1979 when it was found that RNA polymerase II is able to initiate transcription only if additional proteins are added to the reaction. In contrast with the bacterial sigma factors, transcription in eukaryotic cells requires distinct initiation factors that were not associated with the polymerase.
Specific proteins acting as transcription factors have now been identified that are required by RNA polymerase II to initiate transcription. Two types of transcription factors have been defined: general transcription factors involved in transcription from all polymerase II promoters; additional transcription factors involved in control of expression of individual genes.
Experiments using in vitro systems have indicated that five general transcription factors are required for initiation of transcription by RNA polymerase II. The promoters of many genes transcribed by polymerase II contain a sequence similar to TATAA 25 to 30 nucleotides upstream of the transcription start site.
This sequence referred to as the TATA box is similar to the -10 sequence of bacterial promoters and is involved in initiation of transcription as follows: first, a general transcription factor called TFIID (TF indicates transcription factor, II denotes polymerase II) binds to the TATA box. TFIID has multiple subunits including the TATA-binding protein (TBP).
The TBP binds specifically to the TATAA consensus sequence and 10-12 other polypeptides called TBP-associated factors (TAFs). Second, TBP binds to a second general transcription factor (TFIIB) forming a TBP-TFIIB complex at the promoter. Following recruitment of RNA polymerase II to the promoter, two additional factors (TFIIE and TFIIH) are required for initiation of transcription.
Two subunits of TFIIH are helicase that unwind DNA around initiation site, while another subunit is a protein kinase that phosphorylates repeated sequences in the largest subunit of RNA polymerase II. In spite of the development of in vitro systems, much remains to be elucidated about polymerase II transcription in eukaryotic cells.
Transcription by RNA Polymerases I and III:
Like RNA polymerase II, the other two polymerases I and III also require additional transcription factors to associate with appropriate promoter sequences. Although the three eukaryotic polymerases recognise distinct types of promoters, a common transcription factor, the TATA- binding protein (TBP) seems to be required for initiation of transcription by all 3 polymerases.
RNA polymerase I transcribes ribosomal RNA genes which are present in tandem repeats, to yield a large 45S pre-rRNA, which is then processed to derive the 28S, 18S and 5.8S rRNAs (Fig. 15.9).
The promoter of rRNA genes consists of 150 base pairs just upstream of the transcription initiation site. These promoter sequences are recognised by two transcription factors, UBF (upstream binding factor) and SL1 (selectivity factor 1) which bind to the promoter and then recruit polymerase I to form an initiation complex.
One of the four protein subunits of the SL1 transcription factor is TBP. Thus, TBP is a common transcription factor required by all 3 types of eukaryotic RNA polymerases. The promoter of ribosomal RNA genes does not contain TATA box, therefore, TBP does not bind to specific promoter sequences. Thus TBP associates with ribosomal RNA genes through the binding of other proteins in the SL1 complex to the promoter.
The genes for tRNAs, 5S rRNA and some of the small RNAs involved in splicing and protein transport are transcribed by Polymerase III. These genes are characterised by promoters that lie within, and not upstream of the transcribed sequence.
The process of termination of transcription was found out from experiments in prokaryotes in which nucleus is not bound by a membrane. Therefore, synthesis of proteins on ribosomes occurs simultaneously with synthesis of mRNA on DNA.
Visualisation of Transcription:
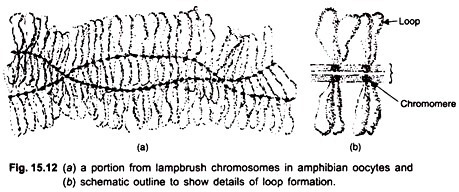
The idea of visualizing gene transcription originally arose from light microscopic studies of lampbrush chromosomes in amphibian oocytes performed by Callan and Lloyd, and Gall in the 1960’s; similar studies were done on the puffed polytene chromosomes of insects by Beermann and his colleagues. Oocyte-chromosomes are highly extended in the lampbrush state and contain thousands of chromosomal loci active in RNA synthesis.
Another favourable attribute of oocytes is that there is amplification (manifold increase) of rRNA genes during early oogenesis giving rise to hundreds of extra nucleoli in a nucleolus. In this system, transcription of ribosomal cistrons has been visualised.
During transcription on oocyte lampbrush chromosomes, the DNA in the condensed, beadlike chromomere unravels and is spun out into a loop and transcribed. The loop axis becomes covered by the transcribed RNA fibrils embedded in a protein matrix (Fig. 15.12). At the base of each RNP (ribonucleoprotein) fibril, an RNA polymerase molecule is attached. In male meiosis and somatic cells transcription produces fine hair-like outgrowths from the chromosomes (Fig. 15.13).
Puffing in the giant polytene chromosomes in the salivary glands of insects represents a direct way of correlating chromosome structure with gene transcription. Puff formation indicates genes that are actively transcribing RNA which would be translated into salivary proteins. Further work of Grossbach (1969) and others made it possible to relate the syn thesis of a specific protein with a specific puff.
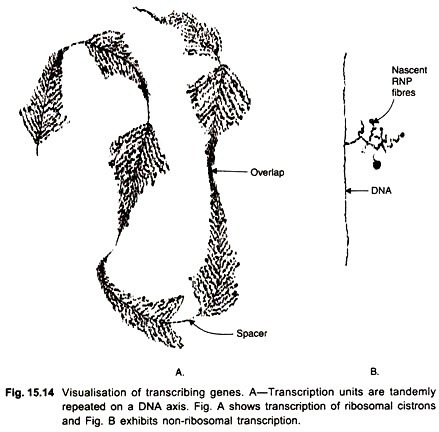
In 1969 Miller and Beatty developed a spreading technique for chromatin by which nascent RNP transcripts could be visualised in the electron microscope. The technique has since been applied to various materials. It allows us to visualize the spatial relationships between DNA, RNA polymerase and the RNA transcripts in situ.
EM studies have confirmed that most of the DNP (de-oxy-ribonucleoprotein) fibrils are entangled in the chromomeric mass and only a small proportion is extended into lateral loops. The initiation and termination sites for transcription are located at the two ends of the loop i.e., the thick and thin insertion sites of the loop.
The lateral RNP fibrils which contain the nascent RNA transcripts are of increasing length. One loop is said to represent one transcriptional unit. Many times an active loop shows tandemly arranged transcription units separated by spacer regions (Fig. 15.14). The spacers represent non-transcribed regions. Up to 5 transcriptional units may be present on a loop; the loop is therefore a multi-gene structure.
These authors also found initiation sites of two transcriptional units overlapping each other. The drug actinomycin-D which inhibits transcription removes RNP fibrils from the template and causes loops to collapse.
The initial products of transcription observed in EM are longer than the average hnRNA molecules isolated by biochemical techniques. Similar studies on transcription have also been conducted on interphase nuclei of somatic cells in some organisms.
Inhibitors of Transcription:
Several compounds can inhibit transcription of DNA by RNA polymerase. One group of compounds acts by binding non-covalently to the DNA template and modifying its structure; the other group binds to the RNA polymerase and inhibits its catalytic function. The most important inhibitor is actinomycin-D (AMD), an antibiotic produced by streptomyces.
Its phenoxazene ring intercalates between two GC pairs, while its two peptide side chains form H-bonds with guanine bases and project into minor groove of the double helix.
AMD does not interfere with the binding of RNA polymerase to DNA but inhibits chain elongation by preventing movement of core enzyme along the template. AMD does not interfere with replication of DNA. Aflatoxin, ethidium bromide and 2- acetyl-amino-fluorine also inhibit transcription by binding to DNA.
A group of bacterial antibiotics called rifamycins act by inhibiting bacterial RNA polymerases. One such compound known as rifampicin binds non-covalently to the β subunit of RNA polymerase so that chain initiation is inhibited, but does not affect chain elongation, α- amanitin blocks one of the RNA polymerase enzymes present in eukaryotic cells; but bacterial mitochondrial or chloroplast RNA polymerases are not affected by it.