In this article we will discuss about the introduction and stability of mRNA.
Introduction to Messenger RNA (mRNA):
The existence of messenger RNA (mRNA) playing the intermediate role of RNA in protein synthesis was postulated by Jacob and Monod in 1961. Before that RNA in ribosomes was thought to perform the function of mRNA. The prevailing notion was, one gene – one ribosome – one protein.
In eukaryotes the primary product of transcription consists of large RNA molecules of variable lengths and sedimentation coefficients between 20S and 200S, called pre-mRNA (earlier heterogeneous nuclear RNA, hnRNA).
The complete RNA transcript or pre-mRNA is not translated into protein. Instead, only a fraction of RNA which has coding sequences is spliced off and modified to produce mature mRNA molecules that are transported through nuclear pores in the nuclear membrane to cytoplasmic ribosomes for protein synthesis.
The coding sequences termed exons are interrupted by non-coding sequences, the introns that never leave the nucleus and are not translated. The introns are removed by splicing, that is they are cut, removed and the coding sequences (exons) are sealed with each other. In contrast, the prokaryotic RNA transcript functions directly as mRNA.
The absence of nuclear membrane around chromatinic DNA in prokaryotes allows ribosomes access to newly formed mRNA and translation into protein begins on the nascent RNA chain, while transcription is still in progress (Fig. 15.10). In bacteria genes performing closely related functions are usually clustered together in the genome.
They are transcribed into large sized polycistronic mRNAs which are translated into several polypeptide chains. Each polypeptide chain is synthesised independently from its own distinct initiation site. In contrast, eukaryotic mRNAs are monocistronic, encoding a single polypeptide chain.
Thus, in eukaryotes, pre-mRNA undergoes processing for 3 distinct modifications before export from the nucleus. Processing includes modifications at both ends of the RNA molecule as well as removal of introns. The 5′ end of pre-mRNA is modified by addition of a structure called a 7-methylguanosine cap.
Capping is initiated by the addition of a GTP to the 5′ terminal nucleotide of the pre-mRNA. Methyl groups are then added to this G residue and to the ribose of the 5′ nucleotide of the RNA chain. The components of a cap are a cluster of methyl groups. Quantitative studies suggest that there is about one cap for every 2,200 bases in He La cell mRNA. The 5′ cap aligns eukaryotic mRNAs on the ribosome during translation.
The 3′ end of the eukaryotic mRNA undergoes a processing reaction for addition of a poly- A tail, called polyadenylation. The addition of a poly-A tail is signalled by the hexanucleotide sequence AAUAAA which is located 10 to 30 nucleotides upstream of the site of polyadenylation.
This is the most conserved sequence for polyadenylation in mammalian cells, and other less conserved sequences also signal polyadenylation. These sequences are recognised by a complex of proteins, including a restriction endonuclease enzyme that cuts the RNA chain.
A separate poly-A polymerase adds a poly-A tail of about 200 nucleotides to the transcript. Polyadenylation signals the termination of transcription. Poly-A tails in most eukaryotes regulate translation and contribute to mRNA stability.
The third modification of pre-mRNA is the removal of introns by splicing. Most genes contain multiple introns. In vitro experiments have shown that splicing of pre-mRNAs proceeds in 2 steps (Fig. 15.11).
First, the pre-mRNA is cleaved at the 5′ splice site, and the 5′ end of the intron is joined to an adenine nucleotide within the intron, near its 3′ end. In this step a bond forms between the 5′ end of the intron and the 2′ hydroxyl group of the adenine nucleotide that results in a closed loop structure. The intron is present in the loop.
Second, a cut is formed at the 3′ splice site, and the two exons are joined by a ligase enzyme. The intron is excised in the form of a closed loop (lariat) that is linearized and degraded within the nucleus. Biochemical experiments have demonstrated that splicing takes place in large complexes called spliceosomes, composed of proteins and RNAs.
The spliceosome contains five types of small nuclear RNAs (snRNAs) called U1, U2, U4, U5 and U6. The snRNAs range in size from 50 to 200 nucleotides, and are complexed with six to ten protein molecules to form small nuclear ribonucleoprotein molecules (snRNPs) which play a key role in the splicing process. 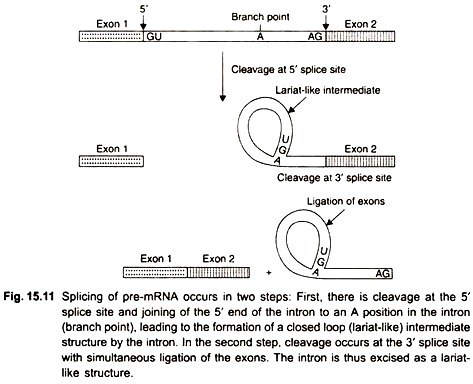
Stability of Messenger RNA (mRNA):
Different mRNAs persist for different time periods in a cell. Since short-lived mRNAs would produce fewer protein molecules as compared with long-lived mRNAs, factors affecting the stability of mRNA determine the level of gene expression. mRNA is degraded by one of two pathways. One route of degradation is the de-adenylation-dependent pathway which begins with enzymatic cutting down of the length of the poly-A tail.
When the poly-A tail is trimmed to a length of 25 to 60 nucleotides, the mRNA becomes susceptible to a de-capping enzyme that removes the 5′ cap so that the molecule is not able to initiate translation. mRNA in this state is rapidly degraded by exonuclease.
The other degradative pathway is the de-adenylation-independent pathway, which is initiated either by de-capping or with endonuclease cleavage of the mRNA, after which exonuclease activity degrades the molecule completely. Thus, the de-adenylation-independent pathway prevents accumulation of truncated (broken) polypeptides in the cell.
The stability of RNA could be affected by a phenomenon known as RNA interference (RNAi) which leads to destruction of RNA transcripts. In this, RNA interference introduces a few hundred nucleotide pairs of double-stranded RNA that triggers degradation of RNA transcripts containing homologous sequences. RNA interference has been seen to occur in a wide variety of organisms including Drosophila, trypanosomes, amphibians, mammals and humans.
It seems that the genes in question are transcribed at the normal level, the transcripts are degraded before they leave the nucleus, and are not translated. This effect of gene silencing is highly specific and requires only a few double-stranded RNA molecules per cell to trigger the silencing. Importantly, the RNAi effect can be transmitted from cell to cell.
