This article throws light upon the three steps of translation process of ribosome.
The three steps of translational process of ribosome are: (1) Initiation (2) Elongation and (3) Termination.
Step # 1. Initiation:
Initiation of translation in E .coli involves the small ribosome subunit, a mRNA molecule, a specific charge initiator tRNA, GTP, Mg++ and number of proteinaceous initiation factors (IFs). These are initially part of the small subunit and are required to enhance binding affinity of the various translational components (Table 8.1). Unlike ribosomal proteins, IFs are released from the ribosome once initiation is completed.
In prokaryotes, the initiation codon of mRNA-AUG- requires modified amino acid, formylmethionine (fmet), in which a formyl group has been added to the methionine’s amino group. The fmet is brought to the ribosome attached to a special tRNA called fmet.tRNA, which has the anticodon 5′-CAU-3′ to bind to the AUG start codon. This tRNA is special, since it is involved specifically with initiation process of protein synthesis.
The amino acylation of tRNA.fmet occurs as follows; Methionyl- tRNA synthetize catalyzes the addition of methionine to tRNA (Fig. 8.3). Then an enzyme called transformylase catalyzes addition of the formyl group to the methionine. The resulting molecule is designated fmet- tRNA.fmet.
Small ribosomal subunits bind to several initiation factors, and this complex in turn binds to mRNA (step I). Protein synthesis is regulated by the sequence and structure of the 5′ un-translated region (UTR) of the mRNA transcript. In prokaryotes, the ribosome binding site (RBS), which promotes efficient and accurate translation of mRNA, is called the Shine-Dalgarno sequence after the scientists that first described it. This purine-rich sequence of 5′ UTR is complementary to the UCCU core sequence of the 3′-end of 16S rRNA (located within the 30S small ribosomal subunit).
Various Shine-Dalgarno sequences have been found in prokaryotic mRNAs (see Figure 8.4 for the consensus sequence). These sequences lie about 10 nucleotides upstream from the AUG start codon. Activity of a RBS can be influenced by the length and nucleotide composition of the spacer separating the RBS and the initiator AUG.
The characteristics of RBS sequences are the following:
1. RBS sequences are rich in purine bases, i.e., rich in Adenine (A) and Guanine (G);
2. They are localized from three to 14 base pairs upstream from the beginning of a gene;
3. Their size varies from three to nine base pairs;
4. Their consensus sequence is “A G G A G”;
5. The RBS sequences are complementary to the pyrimidine-rich sequence found in the rRNA in the 6S unit of the ribosome (end 3′ – HO-AUUCCUCCACUAG-5′).
Initiation of protein synthesis in eukaryotic cytoplasm resembles the process in bacteria, but the order of events is different and the number of accessory factors is greater. Some of the differences in initiation are related to a difference in the way the bacterial 30s and eukaryotic 40S subunits find their binding sites for initiating protein synthesis on mRNA.
In eukaryotes, small subunits first recognize the 5′ end of the mRNA and then move to the initiation site, where they are joined by large subunits (in prokaryotes, small subunits bind directly to the initiation site). In eukaryotes, the Kozak sequence A/GCCACCAUGG, which lies within a short 5′ un-translated region, directs translation of mRNA. An mRNA lacking the Kozak consensus sequence may be translated efficiently in Ambion’s in vitro systems if it possesses a moderately long 5′ UTR that lacks stable secondary structure. Our data demonstrate that in contrast to the E. coli ribosome, which preferentially recognizes the Shine-Dalgarno sequence, eukaryotic ribosomes (such as those found in retic lysate) can efficiently use either the Shine-Dalgarno or the Kozak ribosomal binding sites.
Eukaryotic protein synthesis requires many initiation factors for all stages of initiation, including binding the initiator tRNA, 40S subunit attachment to mRNA, movement along mRNA and joining of 60S subunit.
Another initiation protein then enhances the binding of charged formyl methionyl tRNA to the small subunit in response to the AUG triplet (step II). This step establishes the reading frame so that all subsequent groups of three ribonucleotides are translated accurately. The aggregate represents the initiation complex, which then combines with the large ribosomal subunit. In this process, a molecule of GTP is hydrolysed providing required energy and the initiation factors are released (step III).
Step # 2. Elongation:
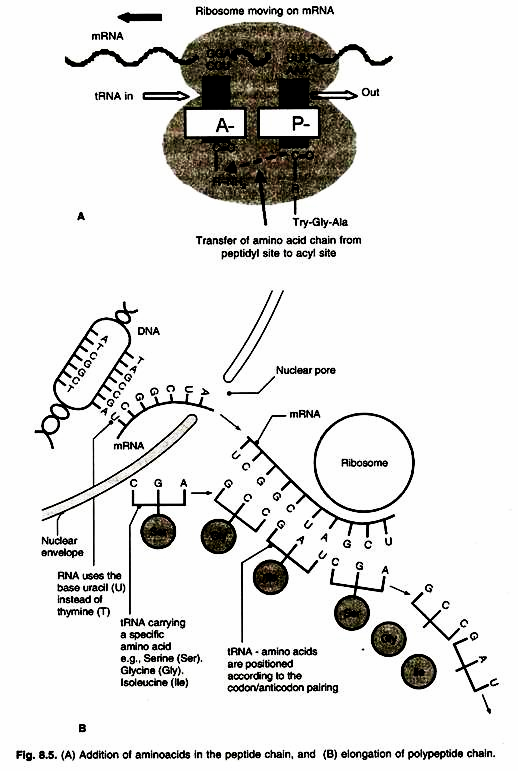
Once both subunits of the ribosome are assembled with the mRNA, binding site for two charged tRNA molecules are formed. These are designated as the ‘P’ or peptidyl and the ‘A’ or aminoacyl sites. The charged initiator tRNA binds to the P site, provided that the AUG triplet of mRNA is in the corresponding position of the small subunit. The increase of the growing polypeptide chain by one amino acid is called elongation.
The sequence of the second triplet in mRNA dictates which charged tRNA molecule will become positioned at the A site (step I). Once it is present, peptidyl transferase catalyses the formation of the peptide bond that links the two amino acids together (stepll). The catalytic activity of peptidyl transferase is a function of rRNA of the large subunit, not one of the ribosomal proteins.
At the same time, the covalent bond between the amino acid and the tRNA occupying the P site is hydrolysed (broken). The product of this reaction is a dipeptide, which is attached to the 3′ end of tRNA still present at the A site (Fig. 8.5 A, B).
Before addition of another amino acid, the tRNA attached to the P site which is now uncharged, must be released from the large subunit. The uncharged tRNA moves transiently through a third site on the ribosome called ‘E’ site (E stands for exit). The entire mRNA-tRNA- aa2-aa1 complex then shifts in the direction of P site by a distance of three nucleotides (step III). This event requires several protein elongation factors as well as the energy derived from hydrolysis of GTP. The result is that the third triplet of mRNA is now in a position to accept another specific charged tRNA into the A site (step IV).
The sequence of elongation is repeated over and over (step V and VI). An additional amino acid is added to the growing polypeptide chain each time the mRNA advances through the ribosome. Once a polypeptide chain is of reasonable size is assembled (~30 amino acids), it began to emerge from the base of large subunits. A tunnel exists within the large subunit, through which the elongating polypeptide emerges.
The principle role of small subunit during elongation is to decode the triplet present in mRNA while the role of large subunit is peptide bond synthesis. The efficiency of the process is very high with one error in 10-4 amino acids added. An incorrect amino acids may present in one of the 20 polypeptides (500 amino acids in length) synthesized. In E. coli, elongation occurs at a rate of ~15 amino acids per second at 37°C.
Step # 3. Termination:
Termination of protein synthesis is carried out by triplet codes (UAG, UAA, UGA; stop codons) present at site A. These codons do not specify an amino acid, nor do they call for a tRNA in the A site. These codons are called stop codons, termination codons or nonsense codons. The finished polypeptide is still attached to the terminal tRNA at the P site, and the A site is empty.
The termination codon signals the action of GTP-dependent release factor, which cleave the polypeptide chain from the terminal tRNA, releasing it from the translation complex (step I). Once this cleavage occurs, the tRNA is released from the ribosome, which then dissociates into its subunits (step II). If a termination codon appears in the middle of an mRNA molecule as a result of mutation, premature termination of polypeptide occurs.