The following points highlight the three special types of chromosomes. The types are: 1. Polytene Chromsomes or Salivary Gland Chromosomes 2. Lampbrush Chromosomes 3. Prokaryotic Nucleoids.
Type # 1. Polytene Chromsomes or Salivary Gland Chromosomes:
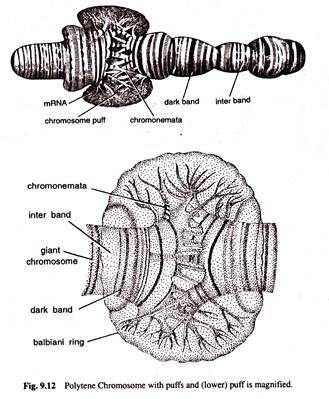
The giant chromosomes were first observed in the cells of salivary glands, gut, trachea and other body parts of dipteran insects by E.G. Balbiani in 1881. The name polytene was assigned to these chromosomes by Kollar. The polytene chromosomes are much larger than the normal somatic chromosomes. How this increase in size of chromosomes is brought about is not known.
The giant chromosomes consist of a bundle of chromonemal fibrils which arise by a series of about 10 consecutive duplications of the initial chromonemata that increase the DNA content about 1,000 times the DNA content of somatic cells (Fig. 9.10). Because of the multi-stranded condition, these chromosomes are called polytene chromosomes.
The process of reduplication of strands without separation is called endoduplication. The homologous polytene chromosomes always remain closely paired in mitotic prophase (Fig. 9.11). This is called somatic pairing and these chromosomes are thought to be in permanent prophase.
The polytene chromosomes bear along their entire length a series of dark bands alternated by light bands or interbands. The dark bands are narrow or broad disc shaped structures. They are euchromatic in nature and contain large amount of DNA, small amount of RNA and certain basic proteins. They are feulgen positive and absorb ultraviolet (UV) light of 2600 A.
The light bands or interbands are fibrillar, feulgen negative, heterochromatic regions containing small amount of DNA. large amount of RNA and acidic proteins and they absorb little amount of UV light.
The number, distribution and localization of discs or bands are notably similar in homologous polytene chromosomes of Drosophila. The centromeres of all these chromosomes fuse to form chromocentre in Drosophila. During certain developmental stages, the single bands or adjacent bands of polytene chromosomes produce local reversible swellings which are called ‘chromosomes puffs’ or bulbs.
The chromonemata of polytenic chromosomes give out many series of loops laterally. These loops or rings are known as the balbiani rings and they are rich in DNA and RNA (Fig. 9.12).
Type # 2. Lampbrush Chromosomes:
Lampbrush chromosomes are special type of giant chromosomes found in the nuclei of oocytes of many vertebrates, such as fishes, amphibians, reptiles and birds during the prolonged diplotene stage of first meiosis. They are also found in the nucleus of Drosophila spermatocyte. Lampbrush chromosomes were first observed by Flemming in 1882 and given the name by Ruckert in 1892.
These chromosomes may sometime become even larger than the polynemic or polytenic chromosomes of salivery glands of dipterans. The largest chromosomes may sometime be as long as 1 mm in urodele amphibians. These chromosomes consist of main axis and many fine lateral projections or loops which give them the appearance of a test tube brush or lampbrush (Fig. 9.13).
Actually, the main axis consists of four chromatids or two bivalent chromosomes and the chromonemeta of these chromatids give out fine loops at the lateral sides. Only the kinetochore bears no lateral loops. Ris (1957) studied the loops with electron microscope and suggested that the loops were integral parts of chromonemata which are extended in the form of major coils (Fig. 9.13).
Generally one to nine loops may arise from a single chromomeral area. The size of loop varies from an average of 9.5µ in frog to nearly 200 µ in newt. These loops probably consist of one DNA double helix from which fibrils project which are covered with loop matrix consisting of RNA and proteins.
Loop formation is interpreted by Gall (1958) as a reversible physiological change which is probably non-genetic. The number of pairs of loops increases in meiosis till it reaches a maximum in diplotene. After diplotene stage, the number of loop pairs gradually decreases and the loops disappear at Metaphase 1.
Physiological studies indicate that the loops of lamp-brush chromosomes and balbiani rings of polytene chromosomes are the sites of active genes.
Type # 3. Prokaryotic Nucleoids:
The cell of prokaryotes or akaryobionta, such as bacteria and blue green algae, are characterized by absence of well organized nuclei. In these cases definite nuclear envelope, as well as nucleioli, is lacking. The electron microscopic studies have revealed that central region of bacterial cell contains feulgen positive granules.
That area has been termed nucleoid or nucleus like structure. Feulgen positive bodies consist of only single circular molecule of DNA which is nearly 1mm long. In cross section, the nucleoid shows 500-900 strands.
This indicates that a single DNA strand in folded forth and back several hundred times. Further work by a number of workers has demonstrated that the network of thread consists of single chromosome in the form of a ring. It is known that the larger part of the DNA in bacterial nucleoid is naked and is not combined with proteins.
The nucleoid of Escherichia coli has a point of contact with plasma membrane. Besides single major circular chromosome, Escherichia coli in fact often possesses one or more minor chromosomes, each called a “plasmid”, which may contain 0.5 to 2% of the total cellular DNA. It is not yet known whether plasmids are located inside or outside the nucleoid.
In blue-green algae, these chromidia are aggregated in the centre of the cell. This also provides clue that the true nucleus with definite nuclear membrane is probably evolved from the primitive type of nucleus.
Chemical Composition of Chromosomes:
The term chromosome refers to deeply stained filamentous body formed of chromatin. The chromatin is composed of DNA, RNA, histones and non-histones.
The purified chromatin isolated from the interphase nuclei shows the following composition:
(i) DNA 30—40%
(ii) RNA 1—10%
(iii) Proteins 50—65%
The chemical composition of chromatin varies considerably from species to species and in different tissues of the same species. The chemical composition of chromosomes also differs at different stages of the cell cycle.
At metaphase chromosomes contain on an average 15-20% DNA, 10-15% RNA and 65-75% proteins. Out of different chemical components of chromatin, DNA is the only component, the amount of which exhibits a constant relationship with the stage of chromosome replication and the increase or decrease in chromosome number.
DNA found in the chromatin may be of two types:
(i) Unique DNA and
(ii) Repetitive DNA.
(i) Unique DNA:
It is DNA consisting of segments whose base sequences are present in a single copy per genome (no repetition in base sequences). Unique DNA segments occur in all the structural genes for histones, ribosomal RNA and storage proteins.
The proportion of unique DNA in genome of different species ranges from 8% (rye) to 70% (man), the average or common values ranging between 40 and 70%. In bacteria the unique DNA represents about 99.7% of the total genome.
(ii) Repetitive DNA.
The DNA in which the base sequences are repeated several to million times per genome is called repetition or repetitive DNA (rep-DNA) by Britten and Kohne (1960). The proportion of repetitive DNA in the genome varies in different species. In man about 30% of genome is represented by repetitive DNA.
Repetition DNA is Characterised by:
(a) The number of nucleotides pairs per sequence,
(b) The specific order of base pairs per sequence, and
(c) The number of copies of a sequence per genome.
A particular family of repeated sequences can exist either dispersed between unique DNA sequences or as clusters of repeating nucleotides units localised in one or few areas called regional repetition in the genome (Lee Thomas, 1973).
The repetitive DNA sequences are grouped into the following two classes on the basis of number of copies per genome; highly repetitive, moderately repetitive DNA and middle repetitive or intermediate fraction.
1. Highly repetitive DNA:
Such DNAs have base sequences repeated more than 105 and upto several million copies per genome. These may constitute upto 10% of the genome and consist of very short nucleotide sequences (sometimes more than 100 nucleotides long). Highly repetitive DNA sequences differ in density from the remaining DNA and are separated as minor bands through Cs Cl2 density gradient centrifugation (Satellite DNA).
Highly repetitive sequences tend to be associated with constitutive heterochromatic regions of chromosomes particularly in the centromere and chromosome ends. It appears to be transcribed to a very small extent, if at all, within the cell.
The probable important functions of such DNAs are participation in chromosomal organisation, involvement in chromosome pairing during meiosis, involvement in recombination and protection of genes for histones and ribosomal RNA.
2. Moderately repetitive DNA.
Such DNAs consist of base sequences repeated 102 to 104 times per genome, each base sequence having 300 ± 200 base pairs. They are interspersed in the majority of instances between the regions of unique or non-rep-DNA and may constitute. 20-80% of the genome.
In man, repetitive DNA fraction is about 13% of the genome. In some eukaryotes this class of DNA may be found in heterogenous state as it may contain many different sequences with different degrees of repetitions.
Intermediate repetitive DNA:
Intermediate fraction transcribed at least in part, includes sequences complimentary to the principal ribosomal RNA (100-500 copies), transfer RNA (16-100 copies) of each species and 5 S rRNA.
In Eukaryotic chromosomes, alternating interspersion of rep-DNA and unique DNA sequences encoding the protein structure shows a high degree of order. The function of most rep-DNA sequences, besides those coding for rRNA, tRNA, 5-S RNA and histones, is presently an open question. Probably rep-DNA represents excess baggages of genome (junks).
They do not transcribe but keep the chromosomes in shape (house-keeping). They serve as gene spacers. They are the material for creating new genes. Regulation of genetic transcription is an another function of rep DNA.