The following points highlight the five types of chromosomes. The types are:- 1. Eukaryotic Chromosomes 2. Bacterial Chromosomes 3. Viral Chromosomes 4. Giant Chromosomes and 5. Polytene Chromosomes.
Contents
Chromosome Type # 1. Eukaryotic Chromosomes:
In 1879 Flemming used the word chromatin for the rod like bodies in the nucleus and Waldeyer coined the term chromosome for the darkly staining bodies in the cell nucleus. Chromo means coloured and soma means bodies.
Sutton and Boveri (1902) suggested that these bodies are carriers of hereditary particles. Morgan (1933) discovered the role of chromosomes in the transmission of hereditary characters.
Chromosomes appear distinct during cell division and have hidden within them all the secrets of genetic characters, which are passed from one generation to the next generation.
The Interphase Nucleus:
In eukaryotic cells, the chromosomes having DNA are present in the nucleus. The DNA contains genes.
During the period between two divisions or the interphase, the chromosomes are in the form of a complex network formed with proteins. The network is called chromatin. The positive charge of proteins neutralizes the negative charge of nucleic acids.
DNA is the genetic constituent of the cells carrying information in a coded form from cell to cell and from one generation of the organism to the next generation. Chromosome is a compact and condensed form of DNA. DNA packed into chromosomes can be transmitted accurately to both daughter cells at the time of cell division.
Fixed Number of Chromosomes:
Every species has a characteristic fixed number of chromosomes. Majority of the eukaryotic organisms are diploid. Their diploid cells possess two copies of each chromosome derived from each parent. The two chromosomes are homologous. Polyploid cells have more than two corresponding copies of chromosomes.
Constant DNA or C-Value:
During 1940s Mirsky, Hans Ris and others proved that the amount of DNA per nucleus is constant in all body cells of an organism. This was a milestone discovery and it suggested that DNA is the genetic material, which carries genetic information.
All the diploid cells of an organism possess the same constant amount of DNA called 2C. Haploid gametes contain half the amount of DNA called 1C.
Constitution of Chromatin:
Chromatin is a complex of DNA and histone proteins. Chromatin consists of DNA, histone proteins, non-histone proteins, RNA, enzymes and metallic ions. Histone proteins and DNA are always present in the ratio of 1: 1. The RNA and non-histone proteins vary in chromatin of various tissues. Enzymes include DNA polymerase and RNA polymerase.
Histone proteins present are of five types — H1, H2a, H2b, H3 and H4. They are rich in basic amino acids like lysine and arginine. Positively charged R group of amino acids of proteins binds to the negatively charged phosphate group of DNA in the ratio of 1: 1 to form deoxyribonucleic proteins (DNP). There are 5000 types of non-histone proteins.
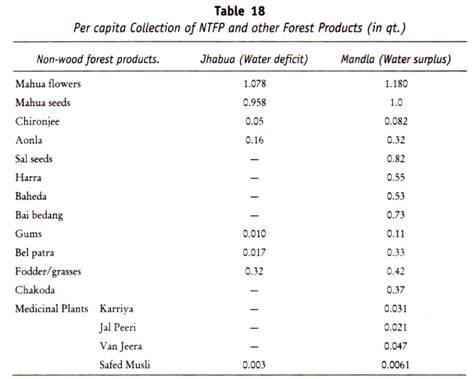
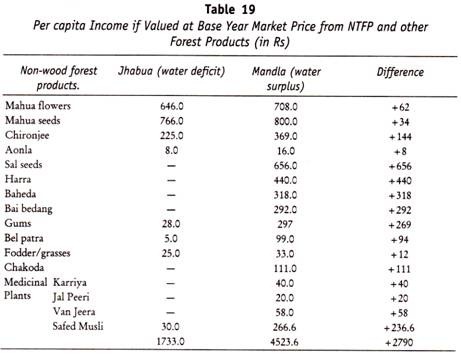
Folding and Condensation of Chromosome:
The human being process 3 x 109 base pairs in the haploid genome. The thickness of each base pair is 3.4Å. If all the DNA molecules were placed end to end, the total length of DNA in a haploid set of chromosomes will be about 10I0A or about one metre. For a diploid set it will be two metre. As the diameter of the human cell nucleus is only 10-15 µm. Only highly folded, compact and condensed DNA can fit into it.
During mitotic cell division, the DNA molecules are highly condensed, contracted, compact, folded and packed. The metaphase chromosomes are folded and condensed about 7000 to 10000 times as compared to the free DNA. On the other hand, during interphase, the DNA is a loose, uncondensed network called chromatin. During different phases of cell cycle, the chromosome undergoes condensation and decondensation cycle.
Chromatin has a Beaded Structure:
In 1973 Woodcock suggested that chromatin is a string of beads made up of repeating units. Under electron microscope, the chromatin has a repeating structure of beads which are about 11 nm in diameter connected by a string of DNA. These beads are called nucleosomes. Association of DNA with proteins forming nucleosomes leads to the folding of DNA. The 11 nm fibre represents the first level of packing and is called beads on a string model.
Nucleosomes:
Four kinds of histones form a bead. Each bead called nucleosome is an octomer containing eight molecules of two proteins each of four kinds. They are H2A, H2B, H3 and H4. Nucelosome is a disc shaped structure of 11 nm diameter. DNA is wrapped around the nucleosome making two turns around each histone octomer.
There are 146 base pairs of DNA in these two turns. Each nucleosome is connected to the next nucleosome by a linker DNA having about 60 base pairs. Thus nuclesome bead and linker DNA have about 200 base pairs.
When nucleosomes are closely applied in 11 nm fibres, the packing of DNA is about 5-7 fold. Thus it is 5-7 times more compact than free DNA.
The 30 nm Fibre:
Nucleosomes are the fundamental building blocks of DNA. These are further folded to form 30 nm fibres.The 30 nm fibres are chromatin fibres of the interphase. Folding of nucleosome chain into a closely packed coiled structure called solenoid structure. In the solenoid model, the nucleosomal DNA forms a super helix structure containing six nucleosomes per turn.
Nucleosome-solenoid model was proposed by Arthur Korenberg and Thomas in 1974. DNA of a 30 nm has a packing of about 40 fold.
During interphase 30 nm fibres form radial loops. This thread containing radial loops is further folded into 300 nm configuration. Most of the chromosome DNA is found in 300 nm configuration and constitutes euchromatin. The 300 nm fibre is further folded into 700 nm fibres which constitute heterochromatin. During metaphase the chromosome having two parallel chromatids has a diameter of 1400 nm.
In highly compact metaphase chromosome, the radial loops remain attached around central scaffold made up of non-histone proteins. Each radial loop has about 5000-88000 base pairs. Two proteins which contribute to the condensation process are condensin and cohesin.
In eukaryotes, as the cell prepares itself to divide, the chromatin becomes highly folded, compact and condensed into individual chromosomes. This helps in their proper sorting and distribution during cell division. These individual chromosomes are visible under optical microscope.
Cell Cycle:
The shape and size of the chromosomes undergoes considerable change during different phases of cell cycle.
The growing cell undergoes a cell cycle that consists of two periods:
1. Interphase
2. Period of Division
In eukaryotes cell division takes place by mitosis. Cells spend most of their life span in interphase, which is the period of intense biosynthetic activity, as DNA synthesis occurs in this phase.
In this phase, the chromosome is completely dispersed and un-contracted. The cell doubles in size and chromosomes duplicate in an exact manner. In this way, before the cell divides by mitosis, chromosomes have already duplicated. Each chromosome has two chromatids. The cell division brings about the final separation of already duplicated chromosomes into daughter cells.
1. G1-chromosomes are completely dispersed and uncondensed.
2. S-Synthesis of DNA-duplication of chromosomes occurs.
3. G2-condensation starts.
4. M-Metaphase. Maximum condensation.
5. A-Anaphase. Two chromosomes formed by division of contromere, go.to opposite poles.
Shape of the Chromosomes:
At the time of cell division, the chromatin becomes condensed into chromosomes. Shape and size of the chromosomes show cyclic changes during different phases of cell cycle. In metaphase of mitotic cell division, they become short, thick, rod like structures.
Each chromosome has a definite shape and size. They are attached to the spindle by means of centromere. The position of the centromere in the chromosome determines its shape.
They are of following types:
1. Metacentric:
Centromeres lies in the centre of the chromosome. They are V-shaped and two arms are equal in length.
2. Sub-metacentric:
Chromosomes have arms of unequal length as the centromere lies near the centre. They are J-shaped or L-shaped.
3. Acrocentric:
Centromeres lie near one end and have a small arm beyond centromere.
4. Telocentric:
The chromosomes have centromeres located on one end on a rod like chromosomes.
Centromere lies in the primary constriction. The region on either side of centromere is dense heterochromatin and has highly repetitive DNA called satellite DNA.
Chromatids:
In metaphase, each chromosome consists of two symmetrical, identical threads called chromatids. Each chromatid contains a single DNA molecule, the unineme theory. This is proved by the fact that G1 chromosome has one chromatid and after replication of DNA, it has two chromatids in G2.
The two sister chromatids are mirror images of each other. The chromatids are attached by centromere. At anaphase each chromatid migrate to the opposite pole, when centromere divides into two. In this way anaphase chromosomes have only one chromatid.
Interphase Chromatin:
This has two types of regions called heterochromatin and euchromatin.
Heterochromatin and Euchromatin:
Some regions of chromatin are densely packed. These regions are caller1 heterochromatin. Some regions of heterochromatin aggregate into dense chromocentre.
It can be darkly strained or false nucleoli. But most regions are much less densely packed. These are called euchromatin. It occupies most part of the nucleus.
The genetic material is organized in a manner that permits alternate states to be maintained side by side in the chromatin. This allows cyclic changes in the packaging of chromatin between interphase and cell division.
The change in structure of genetic material is correlated with its transcriptional activity. Chromatin is not expressed in more condensed state. During mitosis the transcriptional activity comes to a stand still. The heterochromatin is genetically inert as transcription proteins are unable to gain access to the genes whereas the euchromatin is genetically active.
Heterochromatin is of two types:
1. Constitutive Heterochromatin:
Constitutive heterochromatin is permanently condensed. It is most common type of heterochromatin. This includes satellite DNA around the centromere.
2. Facultative Heterochromatin:
It is condensed in certain cells or in certain stage of development. Often one chromosome of homologous pair is condensed. In female mammals one X-chromsome is active and euchromatin while the other is inactive and is carried in the form of heterochromatin state.
Heterochromatin region is the last to replicate. This region is generally inactive and genes in this region are not expressed as they do not synthesize RNA and are switched off.
The switching off of the genes plays important role in cell differentiation.
Centromere:
Centromere is present in the primary constriction and occupies a fixed position on the chromosome, centromeres provide mobility to the chromosome during mitosis.
During anaphase of mitosis, the centromere divides into two. The sister chromatids become separate chromosomes. The centromeres of chromosomes attach to the microtubules of the spindle. Centromeres act as handles for the movement of chromosomes into daughter cells.
They are required for the correct segregation of chromosomes after DNA replication. The two separated chromosomes are called daughter chromosomes. Each daughter chromosome goes to the two daughter cells formed.
Centromere synthesize a fibrous structure called kinetochore to which microtubules of spindle are attached. Most of chromosomes have only one kinetochore.
Centromeres have a definite sequence of bases. In yeast S. cerevisiae, a centromere has a sequence of about 120 base pairs. But majority of eukaryotic centromeres are much longer. Base sequence is A-T rich.
Secondary Constriction:
In addition to the primary constriction, some chromosomes have a secondary constriction. Secondary constriction lies in a fixed position on the chromosome and helps in identifying the particular chromosome. Five human chromosomes have secondary constriction. They are 13th, 14th, 15th, 21st and 22nd. The part of the chromosome beyond the secondary constriction is called satellite and the chromosome is called Sat-chromosome. This satellite is different from satellite DNA.
Secondary constriction has genes for 18 S and 28 S rRNA that induce the formation of nucleoli and are, therefore, called nucleolar organizer region (NOR). Secondary constriction arises because rRNA genes are transcribed very actively.
Telomeres:
Telomeres are tips of chromosomes. Each chromatid has its own telomere. Removal of telomeres makes the chromosomes unstable. Substantial portion of the telomere is single stranded. Their sequence is G-T rich. The G-rich strand protects the terminal end of DNA by forming a cap.
Length of telomere decreases with every cell division and cells are pre-programmed to die (apoptosis). Cancer cells produce telomerase enzyme which prevents telomere shortening and jells become immortal.
Karyotype:
The chromosomal complement of any cell seen at the time of mitosis is called Karyotype. Karyotype reveals many characteristics that help in identification of a particular chromosome set. It includes the number of chromosomes, their relative size, position of the centromere, length of arms, secondary constriction and satellites.
Karyotype diagram represents homologous chromosomes in decreasing size. Mouse has ill acrocentric chromosomes. Banding techniques have helped in identifying the parts of chromosomes.
In 1956 J.H. Tjio and A. Levan discovered the correct number of human chromosomes as 46. In 1960 the scientists at a conference in Colorado, Denver classified 22 pairs of autosomes and two sex chromosomes into seven groups.
1. Group A:
This comprises chromosomes 1, 2, 3. They are largest chromosomes with a median centromere and are metacentric.
2. Group B:
It comprises chromosomes 4 and 5 and are next largest chromosomes. They are sub-metacentric.
3. Group C:
It comprises 6-12 chromosomes and X-chromosome. They are of medium size with sub-metacentric centromere.
4. Group D:
It has chromosomes 13, 14, 15. They are shorter than group C. They are acrocentric with satellites.
5. Group E:
It has chromosomes 16, 17, 18 and are short. 16th is metacentric and 17, 18 are sub-metacentric.
6. Group F:
It has chromosomes 19, 20. They are short and metacentric.
7. Group G:
It has small chromosomes 21, 22 and y-chromosome. Chromosomes 21 and 22 has satellites.
Centromeric index is the ratio of lengths of long arm and short arm of the chromosome.
Banding-Techniques:
In 1968 Caspersson discovered chromosome bands by staining the chromosome with fluorescent dyes such as quinacrine and proflavine etc. As a result of this several types of bands have been found in different chromosomes depending upon the use of different dyes and stains.
The major types of bands are Q, G, C, R, F, N and many others. Each band may possess hundreds of genes. Banding patterns reveal the structural details of the chromosomes. Bands provide various features and landmarks for the identification of different chromosomes of a Karyotype.
The small arm of the chromosome is called p arm and long arm of the chromosome is called q arm. Starting from the centromere, each arm is divided into various bands and regions such as 1, 2, 3,4, 5,…. etc. upto the telomere.
Q-Banding:
Treating the chromosome with fluorescent dye quinacrine, the bands formed on the chromosome are called Q-bands.
G-Banding:
Chromosomes stained with gemsa stain produce bands which are called G- Bands. G-Bands are rich in adenine-thymine. G-bands are important tools in the analysis of
mammalian, birds and reptile chromosomes. But plants lack G-Bands, probably due to more compact chromosomes.
Chromosome Type # 2. Bacterial Chromosomes:
Bacteria contain much more DNA than viruses but much less DNA than eukaryotes. Human beings have 700 times more DNA than E. coli, while E. coli, has 100 times more DNA than bacteriophage lambda λ. The chromosome of E. coli cell is a single, double stranded circular molecule containing 4639221 base pairs.
Chromosomes of most of bacteria are circular molecules of DNA. Covalently closed circular DNA is called cccDNA. Chromosome is in the form of a short, single DNA molecule folded into a compact mass, which is called nucleoid. It is a double stranded helix closed in the form of a circle without any free end. Prokaryote DNA does not have histone proteins and free phosphate groups on DNA are not neutralised by basic proteins but by other substances such as prolamines. Eukaryotic chromosomes undergo condensation-decondensation cycle but bacterial DNA does not undergo any such process and remains in fixed folded state.
In E. coli the total length of DNA molecule is about 1100 µ while the diameter of E. coli cell is only 1-2 µ. Therefore the chromosome can exist only in highly folded and supercoiled form.
Folded genome of E. coli has about 50 loops or domains. Each fold further forms supercoils. Supercoils are produced by DNA gyrase enzyme which introduces negative super- coils. DNA loop domains are stabilized by DNA binding proteins HU and H-NS.
This supercoiled nucleoid occupies about one third of volume of the cell. The DNA of bacterial cell is attached to the inner surface of plasma membrane outgrowth called mesosome. It helps in segregation of replicated chromosome as bacteria do not form spindle like eukaryotes. The two strands separate at one point to form a replication bubble. It is bidirectional and replication forks run in both directions from a single origin called Ori C locus.
The two replication forks that initiate at Ori C move around the genome in opposite directions and meet at a point called termination site or ter site.
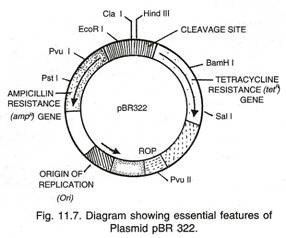
Plasmid:
Plasmid is an additional extra-chromosomal DNA molecule present in many bacteria. Like the main chromosome of the bacteria, they are also double stranded, much smaller and may be circular. They replicate independently of the chromosome.
The plasmids are known to move around freely in the bacterial world. They may pick up genes from one bacterium and transfer them to another bacterium. This may be one of methods of providing variability to the asexually multiplying bacteria.
In some cases, plasmids carry genes that make the host bacteria resistant to antibiotics. Some plasmids carry genes for enzyme β-lactamase, which provide resistance against penicillin and amoxicillin. These plasmids may pass from an antibiotic resistant cell to an antibiotic sensitive cell making the recipient cell antibiotic resistant.
Plasmids are extensively used to clone and modify genes. A modified plasmid is introduced into host cell. Host cell can produce proteins encoded by foreign genes. Example is insulin producing bacteria. Bacteria can survive without plasmids. A plasmid that is able to integrate into host cell’s chromosome is called episome.
Chromosome Type # 3. Viral Chromosomes:
Viruses contain very small genetic information because they use the resources of the host cell for most of the processes required to propagate.
A viral genetic material generally consists of a single molecule of nucleic acid surrounded by a protein coat.
The nucleic acid may be DNA which may be double stranded or sometimes single stranded. Nucleic acid may be RNA which is generally single stranded or sometimes may be double stranded.
The viruses having DNA as the genetic material are called DNA viruses and those having RNA as genetic material are called RNA viruses. In Tobacco Mosaic Virus (TMV), the chromosome consists of a single stranded linear RNA molecule coiled into a spiral. In bacterial viruses bacteriophage the chromosome is double stranded linear DNA molecule.
The viral chromosome has a limited genetic information which codes for little more than production of more virus particles.
Double stranded DNA — Adenovirus, Coliphage T2, T4, T6, PM2.
Single stranded DNA — Phage fd, parvovirus, φ X 174, F1, M13
Single stranded RNA — TMV, Poliovirus, f2, QB.
Double strands RNA — HIV virus, φ6.
In HIV virus, the RNA directs the synthesis of DNA complementary to itself by reverse transcription in the host. The DNA then transcribes RNA for the formation of new virus particles. In TMV the genome is in the form of a coiled helical form of RNA. The protein shell called capsid is made of many small subunits. These subunits are assembled around the helix of RNA. The capsid fits tightly around the genome. Similar spherical capsid is formed around a single stranded RNA in Turnip yellow mosaic virus (TYMV).
The spherical or polygonal capsid of DNA viruses is assembled from protein molecules. Some viruses have genomes that consist of multiple nucleic acids molecules. Reovirus contains ten double stranded RNA segments, all of which are packed into the capsid.
Some plant viruses are multiparticles. Their genomes consist of segments each of which is packaged into a different capsid. Alfa Alfa mosaic virus has four different single stranded RNAs, each packaged independently into a coat. Four components of the virus exist as particles of different sizes.
HIV virus contains about 9000 nucleotides and bacteriophage φβ has 4220 nucleotides both having single stranded RNA genomes.
Bacteriophage φ x 174 has 5386 nucleotides in a single stranded DNA genome.
Chromosome Type # 4. Giant Chromosomes:
Certain special chromosomes are greatly enlarged at a certain stage of their cell cycle. Two examples are Lamp-brush chromosome and Polytene chromosome. They are fully extended chromosomes.
Lamp-brush Chromosomes:
They are found in oocytes of some amphibians in diplotene stage of meiotic prophase during unusually long meiosis. They are the largest known chromosomes. Later they revert back to their normal size. In Lamp-brush form, their length is 400-800 jam while their normal length in contracted form is only 15-20 µm. They were first reported by Flemming in 1882. Ruckert named them and described them in detail.
In meiotic prophase, they are present in the form of bivalents in which the material and paternal chromosomes are held together by Chiasmata. Each bivalent has four chromatids. Lateral loops extrude in pairs from each sister chromatid. Loops are symmetrical, each loop is a single linear thread of DNA that runs through each chromatid. The loops are extruded part of DNA which is being actively transcribed, therefore they are surrounded by matrix of ribounclear proteins (RNP).
Chromosome Type # 5. Polytene Chromosomes:
The salivary gland interphase chromosomes of Drosophilala melanogaster and other dipterans like flies, larvae are greatly enlarged and are called polytene chromosomes.
These multithreaded chromosomes were discovered by Balbiani in 1881 from the salivary glands of chironomus.
The synapsed diploid chromosomal pair undergo repeated replication. The replicated threads are unable to separate as there is no nuclear division. This is called endo-reduplication. The resulting daughter chromatids do not separate and remain side by side attached to the chromocentre, which is formed by the fusion of centromeres.
The number of threads may be in hundreds. After ten rounds of replication, the number of threads is 1024 (210). Each thread represents a single 1C haploid chromosome. Polytene chromosomes are unable to undergo metosis and the cells ultimately die.
Each haploid chromosome consists of series of dark bands consisting of mass of DNA. These bands can be darkly stained by Feulgen dye. In between these bands are lightly stained inter-bands. Some of these bands are further expanded to form reversible chromosomal puffs or Balbiani rings.
These puffs are due to loose coiling of chromatids. These puffs are associated with intense metabolic activity and are sites of active RNA synthesis. Puffs have RNA polymerase II enzyme, which is involved in the process of transcription.
The structure displayed by Lamp-brush and polytene chromosomes suggest that during transcription, the DNA is unpacked and uncondensed from its usual more tightly packed state.
Summary:
Eukaryotic Chromosomes:
Waldeyer coined the term chromosome. In eukaryotic cells, the chromosomes having DNA are present in the nucleus. The DNA contain genes. DNA is the genetic material carrying information in a coded form. The number of chromosomes is fixed for every species. Therefore, the amount of DNA is constant in all body cells of an organism (2C). In the interphase nucleus, DNA and histone proteins are in the ratio of 1:1 and constitute chromatin.
During mitotic cell division, the chromatin becomes highly compact and folded into chromosomes. The metaphase chromosomes are folded and condensed about 7000-10000 times as compared to free DNA. During different phases of cell cycle, the chromosomes undergo condensation and decondensation cycle.
The first level of folding forms a beads on a string model. The beads are called nucleosomes. This bead on a string fibre is 11 nm in diameter. DNA is wrapped around the nucleosomes. Further folding into a solenoid model form 30 nm fibre.
The cell cycle consists of two periods:
1. Interphase,
2. Period of division.
The interphase consists of G1 phase, S-phase (synthesis phase in which amount of synthesized DNA becomes double) and G2-phase. In mitosis, the two chromatids become separate chromosomes and go to different daughter cells. Based on the position of centromeres, the chromosomes are of four types, metacentric, sub-metacentric, acrocentric, and telocentric.
The chromatin has two types of regions, heterochromatin which is dark and condensed region. The second is much less dense called euchromatin, which occupies most part of the nucleus. Centromere is present in primary constriction. Centsomere attach to the spindle and after division into two, facilitates movement of chromosome into daughter cells.
Five human chromosomes have secondary constriction. They are 13th, 14th, 15th, 21st, and 22nd. The portion of chromosomes beyond secondary constriction is called satellite. The tips of chromosomes are called telomeres. The chromosomal complement of any cell at the time of mitosis is called karyotype. Tjio and Levan discovered the correct number of chromosomes in humans as 46.
Bacterial Chromosomes:
Chromosomes of most of bacteria are circular with no free end. Chromosome is in the form of a short, single, double stranded DNA molecule folded into a compact mass called nucleoid. This is further folded into loops, which have supercoils. Most bacteria have an additional extra-chromosomal DNA molecule called plasmid. Plasmid move around freely in the bacterial world and carry genes, which make bacteria resistant to antibiotics.
Viral Chromosomes:
Viruses contain very small genetic information because they use resources of the host cells to propagate. Viral chromosome is a single nucleic acid molecule surrounded by a protein coat. The nucleic acid may be DNA or RNA. The DNA may be double stranded—Adenovirus.
Single stranded DNA virus — Provirus.
Single stranded RNA — TMV, Poliovirus
Double stranded RNA — HIV virus.
Giant chromosomes:
Certain special chromosomes are greatly enlarged at a certain stage of their cell cycle. Lamp-brush chromosomes are found in the oocytes of some amphibians in diplotene stage of meiotic prophase during unusually long meiosis. They are the largest chromosomes known, about 400-800 µm while their normal length in compact form is 15-20 µm. Lateral loops extrude in pairs.
Polytene chromosomes are found in salivary glands of drosophilla. They are multithreaded formed by repeated replication. The chromatids fail to separate. The number of chromatids may be more than a thousand in a single polytene chromosome.