In this article we will discuss about Chromosomes:- 1. Introduction to Chromosomes 2. Chromosome Number 3. Size 4. Morphology 5. Chromosomes in Nucleoprotein 6. Chromosomes Containing Single DNA Molecule 7. Types 8. Models 9. Chemical Structure 10. Biological Importance.
Contents:
- Introduction to Chromosomes
- Chromosome Number
- Chromosome Size
- Morphology of Chromosome
- Chromosomes in Nucleoprotein
- Chromosomes Containing Single DNA Molecule
- Types of Chromosomes
- Models of Chromosome Structure
- Chemical Structure of Chromosomes
- Biological Importance of Chromosome
Contents
- 1. Introduction to Chromosomes:
- 2. Chromosome Number:
- 3. Chromosome Size:
- 4. Morphology of Chromosome:
- 5. Chromosomes in Nucleoprotein:
- 6. Chromosomes Containing Single DNA Molecule:
- 7. Types of Chromosomes:
- 8. Models of Chromosome Structure:
- 9. Chemical Structure of Chromosomes:
- 10. Biological Importance of Chromosome:
1. Introduction to Chromosomes:
A chromosome can be considered a stainable threadlike nuclear component having special organisation, individuality and function. Their presence was first demonstrated in the eukaryotic cell by E. Strasburger in 1875 and these were first termed as chromosomes by W. Waldeyer in 1888.
This term is actually taken from Greek chromasoma which means “coloured bodies” (chroma = colour; soma = body) due to their marked affinity for basic dyes as a consequence of which they axe stained. This property is known as chromaticity.
Staining the cell with certain types of stain (e.g., Aceto-orcein, Acetocamine, Feulgen’s stain) shows that chromosomes are not visible in the interphase nucleus or metabolically active nucleus due to their high water content, but can be easily seen during cell division characteristics whether mitosis or meiosis.
During cell division, the chromosome undergoes dehydration, spiralisation and condensation. So they become progressively thicker and smaller and, accordingly, the satiability of chromosome also increases. Hence the chromosome becomes readily observable under microscope. Staining of chromosomes is generally carried out to make them visible under light microscope.
Chromosomes are capable of duplication and maintaining their morphologic and physiologic properties through successive cell divisions. It has also been demonstrated that the chromosome contains DNA, which in turn, carries the genes and thus plays a major role in heredity.
When reproduction of organism takes place, they are passed on to the next generation through the gametes. Besides, they play an important role in variation, mutation and evolution, and in their control of morphogenesis, multiplication and equilibrium of vital processes.
The term chromosome is mainly used to describe the chromosome of eukaryotic cell. The naked DNA of prokaryotes and DNA or RNA of viruses are sometimes broadly called prokaryotic chromosome and viral chromosome, respectively, due to their similarity in fundamental properties with eukaryotic chromosomes.
But the morphology and the organisation of eukaryotic chromosome is much more complex. The morphology of chromosomes in all eukaryotes is essentially similar—except some variations in number and size.
Most of the chromosomes in an eukaryotic cell are called autosomes which control all somatic characteristic of an organism [These are symbolized by ‘A’]. But, in addition, there are some other chromosomes which control some specialised characteristics of an organism and are called allosomes.
Sex chromosome (X and Y) for determination of sex, B-chromosomes, L-chromosomes, M-chromosomes, S-chromosomes and E- chromosomes are examples of allosomes. Autosomes are universally present in all eukaryotic-organisms, but allosomes may or may not be present in all organisms.
2. Chromosome Number:
The number of chromosomes varies from species to species but it remains constant for a particular species. The number of chromosomes serves as an aid in the determination of phylogenetic status, such as taxonomic position of plant and animal species.
In higher organisms, each somatic cell contains one set of chromosomes inherited from the maternal (female) parent or organ and a comparable set of chromosomes (homologous chromosomes or homologues) from the paternal (male) parent or organ.
The number of chromosomes in this dual set is called the diploid (2n) number. The suffix ‘ploid’ refers to chromosome “sets”. Homologous chromosomes are two copies of a chromosome (one comes from the female and the other from the male parent or organ)—which are ordinarily identical in size and shape, gene content and gene order.
Sex cells or gametes—which contain half the number of chromosome set found in somatic cell—are referred to as haploid cells (n). A genome is a set of chromosomes corresponding to the haploid set of a species. The number of chromosomes in each somatic cell is the same for all members of a given species.
Chromosome number varies widely and may be very low or high in both plant and animals. In animal kingdom Ascaris megalocephala var univalens shows a single pair of chromosomes in the cells of the germ line.
But, since in the diploid soma the two chromosomes split into numerous small chromosomes, the single haploid chromosome has to be considered an aggregate chromosome or compound chromosome. It, for reasons unknown, maintains its unity under the conditions imposed by the cells of the germ line.
Again, the next lowest diploid chromosome number (2n) recorded so far in animals is four in Mesotoma (flat worm) and Ophryotrocha puerilis (Polychaeta). The highest diploid chromosome number (2n) in animals is 254 in Eupagurus schotensis (a hermit crab). Belar (1926) has, in fact, recorded that Aulacantha, a radiolarian has as a diploid number (2n) approximately 1,600 chromosomes.
In plant kingdom, Haplopappus gracilis, a member of the family Asteraceae, has four chromosomes in its somatic tissues which is the lowest chromosome number known in plants. In Ophioglossum reticulatum, a member of eusporangiate primitive fern under pteridophyta, up to 1,260 chromosomes (2n) have been reported. This is the highest chromosome number known in plant.
The somatic chromosome number generally remains constant among individuals of the same species. But in many species, somatic cells of the same individual may exhibit different (2n, 4n, 8n etc.) chromosome numbers.
In such species, cellular differentiation is often accompanied in some cells with a phenomenon of endomitosis. Endomitosis means the duplication of chromosomes without division of the nucleus, resulting in increased chromosome number within a cell. Chromosome strands separate but the cell does not divide.
Endomitosis leads to the production of endopolyploid cells having 2n, 4n, 8n etc. chromosomes. In natural polyploid individuals, it becomes necessary to find out the ancestral chromosome number which is represented by x and is called as the basic number.
For example in common wheat Triticum aestivum 2n = 42; n = 21 and x = 7 showing that common wheat is a hexaploid (2n = 6x). The whole collection of chromosomes in the nucleus of an organism is referred to as chromosome complement.
3. Chromosome Size:
The size of chromosome of a cell shows a remarkable variation depending upon the stage of cell division. Chromosomes are longest and thinnest during interphase. But on the onset of prophase there is a progressive decrease in size associated with an increase in thickness.
Chromosome are smallest during anaphase. But the measurement of chromosome size are practically taken during mitotic metaphase when they are very thick, quite short and well-spread.
The size of mitotic metaphase chromosome of various plants and animals varies from 0.5µ to 32µ in length and 0.2 µ to 3.0 µ in diameter. The giant chromosome found in the cells of salivary gland of Diptera are permanently in pre-metaphase stage and are easily visible in the interphase nucleus. These chromosomes are 300 µ in length and 10 µ in diameter.
Generally, plant chromosomes are longer than animal ones. In angiosperm, chromosomes of monocots are bigger than those of dicots and other plants. The longest metaphase chromosomes of plant are found in Trillium.
The size of each chromosome is 32 µ long. Again, chromosome size of Trillium is hundred times bigger than its closely related genus Medeola, size differences may be seen in the different species of a genus. For instance, the chromosomes of Allium Porum are half the size of the chromosome of Allium sativum.
The size of chromosome may vary in the different tissues within a single organism. For example, in plant Medeola, the root tip chromosomes are 50% bigger than the shoot tip chromosomes.
Among animals, grasshopper, crickets, mantids, newts and salamanders have big chromosomes. In animals, size variation of chromosome has also been reported in different varieties within a species.
For example, the chromosome size of chironomous thumii thumii (fly) differs from its closely related varieties. During embryogenesis of certain marine insects the size of chromosomes of the early blastula are smaller than those of the later stage of development.
The size, shape and number of the metaphase chromosomes constitute the karyotype which is distinctive for each species. When all chromosomes of a species are more or less equal in size, the karyotype is called symmetrical karyotype.
Asymmetrical karyotype refers to the chromosome of different size. In most organisms, all cells have the same karyotype. However, species that appear quite similar can have very different karyotypes—indicating that similar genetic potential can be organised on chromosomes in very different ways.
Variation in the size of the chromosome can be induced by some factors:
i. When the cell divides at low temperature, the size of chromosomes become short and more compact.
ii. When the pre-treatment of cells is done with colchicine, the chromosomes become slightly shorter in size.
iii. Repeated and rapid cell divisions tend to result in smaller chromosomes.
4. Morphology of Chromosome:
It has been observed that the morphology of chromosome changes with the stage of cell division. During the prophase of meiosis, homologous chromosomes pair with each other at zygotene, the cell then enters the stage of pachytene where chromosomes become shortened and coiled.
Pachytene stages are very useful for the study of chromosome morphology because they are longer than the chromosomes in mitotic metaphase, so that the structural details of chromosomes can be easily resolved.
But the meiotic cell division as well as the pachytene stages are not readily available at any time for experimental purpose. On the other hand, mitotic metaphase is easily available by arresting the divisional cycle with some chemical agents. Further, mitotic metaphase is also suitable as convenient stage for studies on chromosome morphology and some of the features are more clear during mitotic metaphase.
At metaphase, each chromosome is made of two symmetrical structures, the chromatids. They are also called sister chromatids. Each chromatid contains a single DNA molecule. The chromatids are held together closely by the centromere (Fig. 13.1) and become separated at the start of anaphase when the sister chromatids move to opposite poles.
Therefore, until two sister chromatids share the common centromere, they are called chromatids. But as soon as they are separated at anaphase and possess their own individual centromere, they are called chromosome instead of chromatid. Hence, from anaphase to next G1 phase, chromosomes have only one chromatid while from S phase to metaphase chromosomes have two (Fig. 13.2).
The DNA present in each chromosome (made of a single chromatid) replicates during S phase to produce an identical copy of itself so that during G2 prophase and metaphase each chromosome is composed of two chromatids.
During prophase, and sometimes during interphase, the chromosomal material becomes visible as very thin threads which are called chromonemata and which represent chromatids in early stages of condensation. ‘Chromatid’ and ‘chromonemata’, therefore, are two names of the same structure. It is now accepted universally that chromatid is the structural and fundamental unit of chromosomes.
The region where two sister chromatids are held together is called the centromere. This region generally appears as a constricted or narrowed zone in the centromere, hence it is also known as primary constriction. Sometimes centromere appears as gap during metaphase because it does not take up any stain.
Centromere has a clear zone in which the fibrils remained uncoiled or less coiled than those in the rest chromosomes. The reduced similarity of centromere” is understandable as the chromosome region in centrosome is less coiled or uncoiled and is composed of heterochromatin.
At or near the centromere of each chromosome, the centromere is associated with a specialised structure called kinetochore. The structure of kinetochore is complex and is seen during late prophase. In ultra-thin sections of chromosomes, the kinetochore is seen as a stack of three-layered proteinaceous disc like plates (Fig. 13.3).
The innermost disk (40-60 nm) probably consists of chromatin that is condensed differentially than the surrounding heterochromatic chromatin. The outer disk is a fibrous structure where kinetochore microtubules are attached by their ends.
A 25 to 30 nm layer—the middle layer separates the inner and outer disk. A series of fine filaments which bridge the middle layer may help to hold the two disks together. The kinetochore microtubules—that extend toward the spindle pole of the cell play an active role in movement of chromosomes toward the poles during anaphase.
The location of centromere and, hence, that of the kinetochore is directly controlled by a unique segment of chromosomal DNA termed centromeric DNA. Much of the information about the structure of auto mere has been obtained from genetic studies of the simple centromere in yeast.
Yeast centromere is very small and binds, a single microtubule. Sequence analysis of cloned centromeric DNAs (CEN DNAs) from yeast chromosomes shows that they are generally organised into three regions—centromeric DNA elements (CDEs) I, II and III (Fig. 13.4).
Out of the three regions CDF II and CDF III appear to mediate interactions with microtubules through centromeric binding factors (CBF) 2 and 3, respectively, and through the kinesin-related protein Kar3p. These proteins are an important link between kinetochore microtubules and the centromere CDE I is conserved in sequence but is not required.
Each chromosome in a genome can be distinguished on the basis of the position of centromere which divides the chromosome into two arms of varying length. The portion of the chromosomes on either side of the centromere is called the arm of chromosomes—which may be equal or unequal.
In case of unequal chromosome arms, one arm of a chromosome is longer than the other, hence they are termed long arm or q arm, and short arm or p arm, respectively.
Depending on the position of the centromere, chromosomes may be divided into four categories:
metacentric,
sub-metacentric,
acrocentric and
telocentric chromosome (Fig. 13.5).
In a metacentric chromosome it occurs at the centre, i.e., the centromere is median so that the two arms of such chromosomes are equal. The metacentric chromosomes look ‘V’-shaped during anaphasic movement. In Trillium and Tradescantia, all the chromosomes are metacentric.
The sub-metacentric chromosomes look ‘L’-shaped in anaphase and the centromere is located on one side of the centred point, i.e., the centromere is sub-median so that it divides the centromere into two unequal arms.
In acrocentric or sub-telocentric or sub-terminad centromere, the centromere is situated almost near one end of the chromosome, i.e., centromere is sub-terminal in position and it gives two arms—one exceptionally short and the other long. Acrocentric chromosomes look ‘J’-shaped in anaphase. Chromosomes of locust and some grasshoppers are acrocentric.
In some chromosomes, however, centromeres appear to be located at one end of the chromosome, i.e., in the position normally occupied by one of the two telomeres. In this type, one arm is more or less equal to the length of the chromosome, and, other arm beyond the centromere is represented simply by dot.
Acrocentric chromosome may appear ‘rod- shaped’ or T-shaped in anaphase. Telocentric chromosomes are of rare occurrence. Telocentric chromosome exist normally in certain species of holomastigote protozoa.
Telocentric chromosomes arising through a transverse fracturing of the centromere are believed to be unstable due to the centromere’s irregular manner of division. This type of division is also known as misdivision of centromere—a process which leads to the formation of iso-chromosome (those in which the two arms are of equal length and are genetically homologous with each other).
Misdivision of centromeres has been observed in Pea, Datura, Wheat and Fritillaria. Telocentric chromosomes have been experimentally produced in wheat, maize etc. Usually, each chromosome has only one centromere but, in some species, each chromosome has more than one centromere. Again, in some cases the centromere is absent.
Hence, depending on the number of centromeres, chromosomes are classified as given:
(a) Acentric:
The chromosome is without any centromere. Acentric chromosome is very rare occurrence. It may arise due to unequal breaking of chromosome arm into two so that only one part has the centromere while, in the other part, centromere is absent.
The part of chromosome having no centromere is called acentric fragment. Due to lack of centromere, acentric chromosomes are not able to attach with spindle fibres and they do not take part in cell division. Ultimately, the cell eliminates the acentric fragments.
(b) Mono-Centric:
Mono-centric chromosomes have only one centromere. It is a very common occurrence in most of the species.
(c) Dicentric:
A chromosome has two centromeres. Dicentric chromosome may be produced as a result of translocation, paracentric inversion etc. If the two centromeres tend to move to opposite poles during anaphase, the chromosome breaks. Rarely a new centromere may appear on the chromosome resulting in an abnormality. Such a centromere is called a neo-centromere. Dicentric chromosome is reported in the cells of wheat.
(d) Polycentric:
In addition to the shapely localised type of centromere described above, there exists a type of non-localised centromere where each chromosome has more than two centromeres. Such chromosomes are called polycentric.
Polycentric chromosomes are found in plant Luzula purpurea (Fam Juncaceae), in the generative tissue of Ascaris megalocephalla univalens, and in Thyanta. In both the above cases, the centromeric property is confined to one or more definite locus of the chromosome so that such centromere is Called localised centromere.
However, in many insects—e.g., most homopteran and hemipteran insects—the centromere activity is non-localised and spread over the entire chromosome length. In such cases, the centromere is called a diffused centromere.
Polycentric chromosome often break into a number of smaller fragments. Each small segment functions independently. For instance, in case of Ascaris megalocephala univalens, the zygotic cell contains only two chromosomes.
But during embryonic development these chromosomes break in the somatic cells so that the cell may have up to 42 chromosomes. However, the cell that will give rise to the generative cell contains only two chromosomes.
Non-staining gaps are seen in certain chromosomes in addition to the primary constriction regions. These regions are called secondary constrictions (Sc). Generally, secondary constrictions are located in the short arm of chromosomes near end but in many chromosomes they are located in the long arm and sometimes they may be present on both arms.
Secondary constrictions are constant in their position and extent. These constrictions are useful in identifying particular chromosomes in a set.
The number of Sc in a genome varies from species to species. In some species, a somatic cell contains at least a pair of chromosomes with Sc while other chromosomes within the same cell are without Sc. In some other species, the number of chromosomes with Sc may be four (e.g., Vicia hajastana), six, eight, ten (e.g., human somatic cell).
Secondary constrictions are distinguished from primary constriction:
i. Sc is without kinetochore.
ii. Sc is not able to attach with spindle fibre during anaphasic movement.
iii. Sc shows the absence of marked angular deviation of the chromosomal segments during anaphase.
Certain Sc contains the genes coding for 18S and 28S ribosomal RNA and that induce the formation of nucleoli. Since they are usually sites for the organisation of the nucleolus they are also called nucleolus organising regions or NOR.
The region of the chromosome separated from the rest of the chromosomes by NOR or Sc is a rounded body called satellite or trabant. If a fine thread is seen between satellite and the rest of chromosome, it is the satellite stalk and the chromosome is a SAT-chromosome.
The satellite and the constriction are constant in shape and size for each particular chromosome. In man, the nucleolar organizers are located in the secondary constrictions of chromosomes 13,14,15, 21 and 22—all of which are acrocentrics and have satellites.
The eukaryotic chromosome terminates at either end in a structure called the telomere. Telomeres have special properties when chromosomes are broken, the free ends without telomeres become “sticky” and tend to fuse with other broken chromosomes. However, the intact ends of unbroken or normal chromosomes are stable and show no tendency to fuse with other chromosomes or other ends.
The telomere differs in structure and composition from the rest of the chromosome. Telomere structure has been studied in a number of organisms. All telomeres so far studied have multiple (30-70) repeats of short species-specific sequences such as TTAGGG in humans, in Tetrahymena thermophila, TTTAGGG in plant Arabidopsis thaliana etc.
Although these sequences are. somewhat variable in different species the basic repeat unit in all species has the pattern 5’Ti_4 A0–1G1_83’Telomeres have either two strands of DNA covalently linked with a sub-teiminal nick or have a single- stranded 3′ end, i.e., the G-rich strand extends by 12-16 nucleotides beyond the C-rich strand. The protruding single-stranded DNA portion is also known as DNA primer.
Telomere sequences are added by a special enzyme called a telomere terminal transferase or telomerases. Telomerase is a ribonucleoprotein. It contains a short RNA component, 159 bases long in Tetrahymena, 192 bases long in Euplotes. This RNA provides the template for synthesizing the G-rich repeating sequence to which it is complementary.
Three nucleotides (say … A AC …) of RNA template within the enzyme telomerase pairs with 3′ terminal three nucleotides (… TTG) of DNA primer, as shown in Fig. 13.6. RNA template directs and adds nucleotides to the 3′ end of DNA primer.
It adds G and T bases one at a time to the primer as directed by the template and polymerisation continues to end of template region. The enzyme moves to new 3′ end of template before another round of addition takes place. The cycle starts again when one repeating unit has been added.
The telomerase is a specialised example of a reverse transcriptase, an enzyme that synthesizes a DNA sequence using an RNA template. The added G-rich sequence can fold back on itself to form a novel hairpin or four stranded DNA loop.
This involves G : G base pairing in which one or two of the G bases has the syn configuration (Fig. 13.7). Although such secondary structures may stabilise the ends of the chromosomes, they may also interfere with telomerase action.
The unique structure of telomere of eukaryotic chromosomes performs three important functions:
i. It prevents exonucleases from degrading the ends of the linear DNA molecules.
ii. It facilitates replication of the ends of the DNA molecules without loss of termini.
The unusual behaviour of telomeric fractions are:
i. It consists of a basic repeat unit having the general pattern 5′ T1_4 A0_1 G1_8 3′.
ii. In some species the telomeres terminate with a single-stranded G-rich region of the DNA strand with the 3′ end (a so-called 3′ overhang).
iii. The enzyme telomerase conditions the RNA component which provides the template for synthesizing the G-rich strand. This enzyme acts as a specialised Reverse transcriptase.
iv. The terminal DNA is folded and forms either a duplex hairpin by G-G pairing or a G quartet when one G is contributed by each of the 4 repeating units.
v. DNA polymerases are not able to replicate the terminal segment.
In the meiotic prophase and early mitotic prophase, the chromatin material is seen to have dense thickened areas at regular intervals giving the appearance of a string of beads. These bead-like areas are known as chromo- meres.
The distribution of chromomeres in a chromosome is highly characteristic and constant. Homologous chromosomes show an identical pattern. Chromomeres are specially obvious in polytene chromosomes where they become aligned laterally constituting the chromosome bands.
Chromomeres of a single chromosome show a considerable variation in size. Once it was believed that genes were located within chromomeres and one chromosome may possibly represent a single gene.
But this idea has been a controversial one and available cytological evidence does not support this view. The chromomere can best be considered as a unit of functional coordination. Chromomere represents simply the tightly coiled or folded regions of DNA than in the neighbouring regions of chromosome called inter-chromomeric region.
So they are visible in the phase of cell division. At metaphase, the chromosome is tightly coiled and chromomeres are no longer visible.
5. Chromosomes in Nucleoprotein:
(a) Chromatin:
Eukaryotic chromosomes in metaphase are generally known as chromosomes but in interphase the term chromatin is more generally used to describe the nucleoprotein fibres in the cell nucleus.
On the basis of stain-ability with basic dyes during various stages of cell cycle, chromatin is sub-divided into two main classes:
(i) Euchromatin and
(ii) Heterochromatin.
Interphase chromosome cannot be distinguished individually. The interphase nucleus is seen to contain scattered chromosome material and certain highly condensed bodies or chromo- centre. The euchromatin region takes light stain in the interphase nucleus.
It takes comparatively deep stain during cell division. On the other hand, heterochromatin takes deep stain during interphase and prophase while during metaphase it takes light stain (Fig. 13.8).
The changes of stain-ability can be correlated with the changes of condensation and de-condensation property of the chromosome during various stages of cell division. It is obvious that condensed state takes deep stain while extended form takes light stain.
The distribution of heterochromatin of chromosomes has been studied extensively. Heterochromatin has been found to be located at some specific regions such as centromere, chromomere, nucleolar organising region, satellite etc. (Fig. 13.9). But besides these regions there are some other regions where heterochromatin may be present.
Heterochromatin may be classified into:
(i) Constitutive, and
(ii) Facultative.
Constitutive heterochromatin remains permanently in the heterochromatic state, i.e., it does not turn back to the euchromatic state. It tends to have a constant position on homologous chromosomes, e.g., centromere.
In contrast, facultative heterochromatin is essentially euchromatin that has converted into heterochromatin (heterochromatinisation) whenever it needs and, again, it may be changed into euchromatin state (euchromatinisation).
Facultative heterochromatin is seen to develop during the development of an organism. Heterochromatinisation may involve a segment of a chromosome, a whole chromosomes (for example one X chromosome of human females) or one whole haploid set of chromosome (for example, full male set of chromosomes of mealy bug).
Within a chromosome, constitutive heterochromatin has been found to be located at certain regions:
(i) Centromere,
(ii) Telomere,
(iii) Nucleolus organising regions and
(iv) Intercalary segment of chromosome.
Entire chromosome may show heterochromatic behaviour, e.g., sex chromosomes in some plants, supernumerary chromosomes etc..
Heterochromatin shows different properties. All types of heterochromatin do not exhibit similar properties. Again, all such properties are not associated with a particular heterochromatin. Earlier workers considered that all heterochromatins were largely genetically inactive since addition or loss of heterochromatin did not have an appreciable phenotypic effect.
It has been also suggested that highly condensed heterochromatin is generally not available for transcription and this condition leads to genetic inactivity. But these concepts are not of universal application. Heterochromatin may not be completely devoid of genes or genetic activity.
Muller demonstrated the presence of the “bobbed eye” gene on the Y-chromosome of Drosophila which was heterochromatic. Gates suggested the location of the gene for hairy earlobes in man on the presumed inert Y- chromosome.
In tomato, a gene has been localised on heterochromatic segment. Again, the proportion of genes on heterochromatic and euchromatic regions in salivary gland chromosome was found to be same on a length to length basis.
When a heterochromatin segment is trans-located next to euchromatin segment, the activity of euchromatic genes is heterochromatinised. However, similar properties are seen when euchromatic part is shifted next to heterochromatin. This is called position effect.
Temporary genetic inactivity has been observed in certain chromosome segment, which are facultatively heterochromatic. This property is very useful for explaining the process of differentiation. For example, in certain coccids (insects) a given chromosome becomes heterochromatic during embryonic development and, at the later stage, it turns back to the euchromatic state.
Heterochromatic regions are not able to synthesise RNA in vitro. It indicates that chromatin fibres in heterochromatic segments are more tightly packed than euchromatic regions.
Experiment with the rate of incorporation of tritiated thymidine into different cells at different stages of development reveals that heterochromatic regions are seen to replicate their DNA in synthetic phase later and early than the euchromatic region. This phenomenon can be correlated with transition from euchromatin to heterochromatin.
In certain cases, heterochromatic region contains a high content of repetitive DNA. This explains the low phenotypic effect following the loss or gain of segment. Heterochromatic regions are easily broken by ionising agents and radiomimetic chemicals.
Allocycly or heteropycnosis is another property of heterochromatin. Certain segments of the chromosome are more condensed than the rest of the chromosomes during various stages of the cell cycle. Such differences in thickening has been called heteropycnosis.
It is exhibited principally by the heterochromatin on the two sides of the centromere. This property is not universal because many forms do not show complete allocycly.
(b) Karyotype:
Karyotype represents the chromosome constitution of a cell or an individual. It deals with the length of chromosome, the position of centromere, presence of secondary constriction, and the size of satellite of the somatic chromosome complement. Generally, karyotype is prepared from well-scattered chromosomes of mitotic metaphase plate.
The information regarding chromosome constitutions are obtained from hand drawing of the microscopic view of chromosomes with the help of camera Lucida or drawing prism. Photomicrographs of metaphase plate are also used for the preparation of karyotype.
Karyotypes are presented by arranging the chromosomes in a descending order of length in a straight line. The longest chromosome is always placed on the right side and the smallest one of the right. All chromosomes in a karyotype do not bear the centromere at the same position.
So, in a karyotype, chromosomes are grouped on the basis of the position of centromere and, in each group, descending order of length of chromosome is also maintained. Each chromosome of a karyotype is marked by a serial number from the extreme left to extreme right.
Broadly, karyotypes of different organisms may be classified into two categories:
i. Symmetrical Karyotype;
ii. Asymmetrical Karyotype.
A symmetrical karyotype has all metacentric chromosome of the same length. In case of asymmetrical karyotype (Fig. 13.10) variation of length of chromosome complement is found and the position of centromere may or may not be identical.
In certain case, karyotype is asymmetrical but the length of chromosome is sharply two types—some chromosomes are very long and some are very short. This type of asymmetrical karyotype is known as bimodal karyotype.
It is believed that symmetrical karyotype is the primitive form from which more advanced asymmetrical karyotype has been evolved. The karyotype of a species may be represented on graph or plain paper by bar diagram showing all morphological features of the chromosome. Such diagram is known as Idiotype or Idiogram (Fig. 13.11).
Idiogram is prepared from haploid chromosome complement of an organism. An idiogram gives the identical information like that of karyotype.
6. Chromosomes Containing Single DNA Molecule:
All DNA viruses and bacterial cells contains a single chromosomal DNA molecules. The general belief is that all eukaryotic chromosomes also contain a single long linear DNA molecule. Extraction of longest DNA experimentally from lower and higher eukaryotes leads to hypothesis that each chromosome contains a single DNA molecule.
For example, physical analysis of the largest DNA molecules extracted from several genetically different species and stains of Drosophila exhibits that they are from 6 x 107 to 1 x 108 base pair long.
These sizes match the DNA content of single stained metaphase chromosomes of Drosophila melanogaster, as measured by the amount of DNA specific stain absorbed. Hence, each chromosome possibly contains a single linear DNA.
The correspondence between the number of chromosomes and the number of DNA molecule per cell has been demonstrated in yeast cells. The length of yeast chromosomal DNA ranges from about 1.5 x 105 or 106 base pairs.
The DNA of yeast chromosome can be separated and individually identified by pulse-field gel electrophoresis. The result of this technique shows that the number of separated DNA molecules equals to the number of chromosome in yeast.
The strongest evidence in the support of unineme (having a single DNA double helix per chromatid) concept is provided by studies on lamp brush chromosome. The loops of a lamp brush chromosome represent single chromatid in a fully extended state.
In electron micrographs of RNAase and protease treated lamp brush chromosomes, the loops have a diameter of 20A which is the diameter of a DNA double helix. Overwhelming evidence form a variety of studies supports the theory that each chromatid contains a single giant DNA molecule.
7. Types of Chromosomes:
In a vast majority of plants and animals, besides autosome, the cells of individual may contain one or more than one chromosome(s). These chromosomes are collectively referred to as allosomes. They differ from autosomes by their specialised functional role.
Autosomes are present in all cells of all organism but the existence of allosomes is not always universal. Allosomes are of different types. All types are not necessarily present in one organism.
(a) Sex Chromosomes:
Chromosomes that are connected with the determination of sex, are called sex chromosomes. There are two types of sex chromosomes; X and Y. X chromosome is found in both males and females although one sex has only one while the other sex have two X-chromosomes.
Y-chromosome occurs only in one of the two sexes of a species, e.g., male fruit fly, human, male mice, some male plants and female birds, reptiles. Y-chromosome contains mostly heterochromatin and only few genes are located in it. On the other hand, X-chromosome is made of euchromatin and many genes are located on it.
(b) B Chromosomes or Super Numerary Chromosome or Accessory Chromosome:
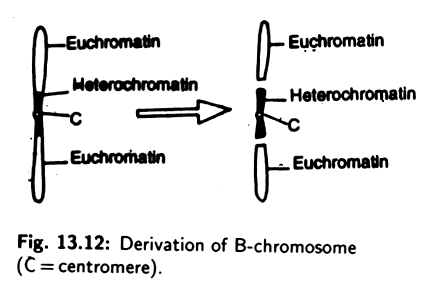
In some organisms, chromosomes in addition to normal autosomes are present as an extra chromosomes that are not genetically necessary for the individual and not homologous to any of the normal chromosomes. These chromosomes differ from the normal ones in their variable number, smaller size and greater degree of heterochromatinisation. They are found more commonly in plant than in animals (Locusta migratoria, Camunla pellucida, Helix pomatia, Myrmeleo tettix maculatus).
B-chromosome do not usually have any effect on the phenotype and, hence, they are not genetically desirable. In some plant their presence results some deleterious effect, i.e., loss of vigour. Though they are found to be deleterious, yet the occurrence of such chromosomes through generations indicates that they must have some positive adoptive role as well.
B-chromosomes do not usually pair with normal chromosome during meiosis though they may pair with each other without the formation of chiasmata when present in even number.
B-chromosomes may be eliminated from certain tissues or organs during embryogenesis.
The origin of B-chromosome is rather obscure. They may have been originated from the ordinary chromosome. It has been suggested that the centric heterochromatin part of an autosome is gradually converted into B-chromosome by the elimination of euchromatin part (Fig. 13.12).
(c) Micro-Chromosome:
These chromosomes are also known as minute or m- chromosomes. They are so-called because of their extremely small, dot like size (about 0.5 µm). Micro-chromosomes are known both in plant (in many species of bryophyte) and animals [in insects of coreidae (Heteroptera), birds etc.].
They have been found mainly during meiosis and occasionally during mitosis. Micro-chromosomes are seen along with large chromosomes or bivalents.
They contain DNA and undergo pairing into bivalents which are sometimes arranged in a rectangle like a quadripartite group. In a peat moss sphagnum there are 19 large bivalents and two m-chromosomes consisting of univalents and four m-chromosomes arranged in quadripartite fashion (Fig. 13.13).
In certain exceptions sat chromosomes are relatively rare and the property of organising nucleoli has been ascribed to specific micro-chromosomes as in bird.
(d) Mega-Chromosomes:
Mega-chromosomes are so called because they are non polytenic and many times longer than the length of normal chromosomes. They are not found in all cells and occur only in a small population of somatic cells. Generally, there is only one mega-chromosome per cell. Sometimes more than one mega-chromosomes have been reported.
Mega-chromosomes may be mono-centric, dicentric or acentric. They are found in the successive generations but they are not transmitted through the gametes. Hence mega-chromosomes are inheritable but the cells are able to produce them. Mega-chromosomes have been reported in a few species of Nicotiana hybrids.
(e) Limited Chromosomes:
Limited chromosomes are large in size and limited in distribution, i.e., they are found only in the germ cells. Limited chromosomes are also known as L-chromosome. They are found in insects of the family Sciaridae (Diptera).
During the embryonic developmental stage particularly the fifth and sixth cleavages limited chromosomes are eliminated from the somatic tissue but are retained in the germ line cells.
Fig. 13.14 shows the schematic representation of L-chromosome in the Sciaridae where the germ line cells of both male and female contain six autosome, sex chromosomes (two X-chromosomes in female and one X-chromosomes in male) and two L-chromosomes.
In somatic cells of both male and female L-chromosomes are absent. Because L-chromosomes are present in all individuals of species in which they are found they are considered to be B-chromosomes.
(f) Somatic Chromosome and Eliminated Chromosome:
Somatic chromosome or S-chromosome and eliminated chromosome or E-chromosome are so called because some chromosomes are retained in both somatic and germ line cells but other chromosomes are eliminated only in somatic cell during early cleavage stages of the embryo.
S and E chromosome have been found in gall insects (fam. Cecidomyiidae) and the insects belonging to the family chironomidiae (Fig. 13.15). In case of Maistor a gall insect both male and females have 48 chromosome in their germ cell and there is no loss of chromosomes. But in somatic cell, 36 chromosomes are lost in female and 42 chromosomes from male.
Hence out of 48 chromosomes, 12 chromosome are present in somatic cell female and 6 chromosomes in male. Chromosomes which axe retained in both germ line cells as well somatic cells are referred to as S-chromosome. Those which are lost or eliminated from the somatic cells but Eire retained in germ cell are known as eliminated chromosome or E-chromosome.
(g) Special Type of Chromosomes/Giant Chromosomes:
In certain eukaryotic organisms there are special tissues where the chromosomes are of special structures not found in other cells of the same organism. These chromosomes attain their largest size in the nuclei of their respective cell. Hence these are also called giant chromosomes.
The giant chromosomes are found in the suspensors of the embryo of certain plant, cells of salivary glands of Drosophila and Chironomous, in the cells of malphigian tubules, epithelium lining of gut of Drosophila, in the cells of fat bodies of larval stage of certain Diptera, oocyte nuclei of certain vertebrate, and invertebrate.
Special types of chromosome have been classified into two categories:
(A) Polytene chromosome and
(B) Lampbrush chromosome.
A. Polytene Chromosome:
Polytene chromosomes are those giant chromosomes in which DNA is replicated in such a way that the daughter chromatids do not separate. In more details, polytene is achieved by replication of the DNA many times without nuclear division (endomitosis) and the resulting daughter chromatids do not separate and remain aligned side by side to form a giant multi-stranded chromosome.
Polytene chromosome first provided the evidence that eukaryotic gene is regulated at the level of RNA synthesis. These chromosomes are the valuable material for the study of gene regulation because their gene transcription can be seen directly in the microscope.
Polytene chromosome differs from polyploidy, in which there is also excess DNA per nucleus, but in which the new chromosomes are separated from each other.
(a) Polytene Chromosomes in Animal Cells:
Polytene chromosome in animal was first observed by E. G. Balbiani in 1881 in the salivary glands of chironomous (a dipteran fly) larva. That is why these chromosomes are also known salivary gland chromosome.
These chromosomes are easy to see in light microscope as large coiled bodies about 150- 200 times as large as gonad cell chromosome. Due to their enormous large size compared to that of normal chromosome they are also called giant chromosomes.
In course of investigations of polytene chromosome in other animal cells it is observed that such chromosomes also exist frequently in the other tissues such as the living cells of the gut, Malpighian tubules muscles, fat cells in some other dipteran like flies, mosquitoes and midges.
The most prominent ones are located in the salivary gland larva of Drosophila melanogaster (fruit fly). These are easily and readily available for cytogenetical study. Hence the salivary gland chromosomes become an ideal material for the purpose of practical as well as research work.
In Drosophila melanogaster these chromosomes—observed in the salivary glands to late larval (3rd instar) stages—are over 100 times the length of the somatic metaphase chromosomes which measure about 7.5 µ. According to Bridges (1938) the salivary gland chromosome of Drosophila measure up to 1180 µ or even up to 2,000 µ.
Those of a related genus Rhyncosciara are even larger and may reach even greater dimensions as a result of pathological disturbances.
The salivary gland chromosomes show somatic pairing at interphase because of their multi-stranded giant nature. Hence the number of these chromosomes in the salivary gland cells always appear to be half of the normal somatic cells (2n = 8).
Another characteristic feature of the polytene chromosome is that along the length of chromosome there is a series of dark bands alternating with other clear zones called inter-bands (Fig. 13.23).
The dark bands are heterochromatic in nature and stain intensely and are Feulgen positive. Furthermore they absorb ultraviolet light at 600A0. There are about 5,000 bands in the Drosophila genome. About 85% of polytene chromosome is in bands and 15% is an inter-bands.
Burke Judd et al have reported about 1,000 bands only on the X-chromosome. The bands of polytene chromosome is thought to represent a looped domain (loops of chromatin that extend at an angle from the main chromosome axis) that is highly folded as shown schematically in Fig. 13.24.
Depending on their size, individual bands are estimated to contain 3,000 to 300,000 nucleotide pairs per chromatin strand. Since the bands can be recognised by their different thickness and spacing’s, each one has been given a number to generate a polytene chromosome “map”.
In polytene cell the chromosomes appear as five long strands and one short strand, attached to a central amorphous mass known as chromo Centre to which the single large nucleolus is attached (Fig. 13.25). The pericentromeric heterochromatin of all the Drosophila chromosomes coalesces in a chromo Centre.
Of the six strands the short one represents chromosome 4 and the larger one represents the X-chromosome while the remaining four are the left and right arms of V shaped chromosome 2 and chromosome 3.
The 4th chromosome, being quite small, is almost completely inserted in chromo Centre and appears as a dot. In female flies, a pair of the X-chromosome appear as a single structure due to somatic pairing. But in male flies ‘X’ chromosome is single. The Y chromosome is fused within the centromere. Hence Y chromosome is not seen as a separate strand.
The DNA in each of the four Drosophila chromosomes has been replicated through 10 cycles without separation of the daughter chromosomes so that 210 = 1024 identical strands of chromatin are lined up side-by-side (Fig. 13.26). Other Dipteran species have more DNA molecules per polytene chromosomes, for example, chironomus has 16,000.
One of the most important characteristics of polytene chromosome is that it is possible to see in them the genetic activity of particular chromosomal site at local enlargements. This is known as puffs or chromosomal puffs or Balbiani rings which are associated with differential gene activation.
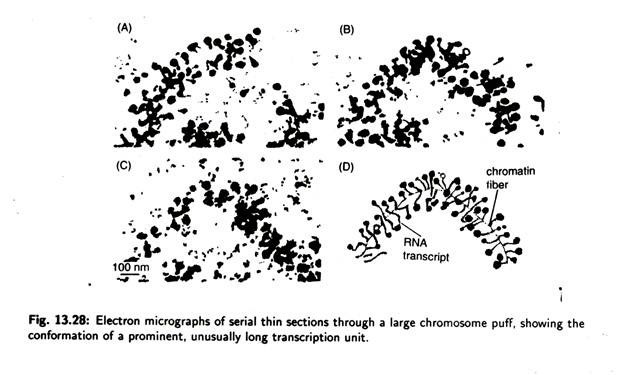
A puff is considered a band in which the DNA unfolds into open loops as a consequence of intense gene transcription, i.e., RNA synthesis. Puffing is a cyclic and reversible phenomenon at definite time and in different tissues of larvae.
Puffs may appear, grow and disappear. Puff formation can be identified by labelling the cell briefly with the radioactive RNA precursor 3H uridine and locating the growing RNA transcripts by autoradiography.
One of the main factors controlling the activity of genes in polytene chromosome is the insects’ steroid hormone ecdysone. During larval development the level of ecdysone goes up and down—that induces the transcription of various gene coding for proteins that the larvae requires for each molt and for pupation.
As the larvae progresses from one developmental stage to another new puff forms and old puff disappears as transcription units are activated and deactivated and different mRNA and proteins are made.
Electron microscopy of thin sections of a puff (Fig. 13.27) shows that the DNA is less condensed. This observations suggest that a looped domain can de-condense as a unit during transcription. It is also observed that puffing can be induced by heat shock when Drosophila larva, normally grown at 25°C, are transferred to a temperature at 37°C, a series of specific gene is activated while most other genes are deactivated.
(b) Polytene Chromosome in Plant Cell:
The study of polytene chromosome in the larvae of Drosophila and other dipteran insects have stimulated investigation of polytene chromosome in other forms of life including plant. In 1962, W. Nagl reported the presence of polytene chromosome in plant. He demonstrated the plant polytene chromosome, in the giant basal cells of suspensors of the embryo of Phaseolus coccineus (common name—runner bean).
Such chromosomes have already been found by Bianchi (1946) in Lloida serotine and later in two plants under Helobie by Hasitschka-Jensche (1955). Unfortunately, Bianchi did not recognize them as polytene chromosome. Hasitschka-Jensche compared them with polytene chromosome of dipteran flies.
H.V. Ivanovaskaya (1973) reported various functional structures in the giant chromosomes occurring in wheat antipodal cells. Similarly, W. Nagl again reported polytene chromosome/nuclei in antipodal cells in Scilla biflora.
Prof. Sima Bhattacharya (1977) reported the presence of polytenic chromosomes in the endopolyploid cell adjacent to the root tip meristem of Phaseolus vulgaris (French bean) Paureus (Mung), Ajuga genevensis.
Polytene chromosomes are bundles of stretched interphase chromosomes which originate by increase in transversal multiplicity as a result of endo-mitotic cell cycle or endo-duplication cycles without separation of the produced sister chromosomes or endo-chromosome.
The types of cell cycles allowing the formation of polytene chromosomes are shown in Fig. 13.28. The length of the giant chromosome is 15 to 30 times greater to that of diploid chromosomes. Both the length and diameter of the polytene chromosome depend on the degree of endopolyploidy.
Plant polytene chromosomes appear to be usually unpaired and therefore present in the diploid number. Nagl (1974) reported the haploid number of polytene chromosomes in antipodal cells. Like polytene chromosomes of Drosophila, plant polytene chromosomes are not showing regular bands and inter-bands. However, the functional units or puffs have been reported by many workers.
DNA replication of polytene chromosomes takes place within 2-4 hours whereas, in metaphase, chromosomes 6-12 are required for the same activity. The other activities of polytene chromosome are RNA synthesis and production of micronucleoli. The significance , of endopolyploidy and polyteny lies possibly in protein synthesis in short-lived but highly specialised cells.
B. Lampbrush Chromosomes in Animal Cell:
Lampbrush chromosome is another type of large diplotene chromosomes present in oocyte nuclei particularly conspicuous in urodele amphibians. These chromosomes have also been reported in the diplotene of oocytes of some fishes, sharks, mollusks, reptiles, birds and spermatocyte nuclei of Drosophila.
Typical lampbrush chromosomes are made of a well- defined chromosomal axis, chromomeres and numerous thin lateral loop extensions. The organisation of the lampbrush chromosome is shown schematically in Fig. 13.29.
Lampbrush chromosomes were first observed by Flemming in 1882 and were described in detail by Ruckert in 1982. The name ‘lamp- brush’ was given because it is similar in appearance to the brushes used to clean the chimneys of oil lamp.
Lampbrush chromosomes have many fine lateral loops, giving them the characteristic ‘hairy’ appearance. Lampbrush chromosomes are found in meiotic prophase, they are present in the form of bivalents in which the homologous chromosomes axe held together by chiasmata. Each bivalent has four chromatids, two in each homologue.
The axis of each homologue consists of a row of granules or chromomeres from each of which one to nine lateral loops may arise. The loops are always symmetrical, each chromosome having two of them—one for each chromatid.
Lampbrush chromosome are up to 300µm long. There are about 10,000 per chromosome set. The size of loops varies from an average 9.5 µ to 200µ. An average sized loop can be estimated to contain roughly, 100,000 nucleotide pair of DNA.
About 5 to 10% of the DNA is present in the lateral loop. The loops may vary in size, thickness and other morphological characteristics. Each loop has an axis formed by a single DNA molecule that is unfolded from the chromosome as a result of intense RNA synthesis.
Lampbrush Chromosome in Plant Cell:
Lampbrush chromosomes have been convincingly demonstrated in some animal cells. But there was no report of such chromosomes present in plant cell.
However, there have been claims in the literature of the occurrence of lampbrush like chromosomes in the diplotene stages of Allium cemum and a fungus—the basidiomycetes coprinus. But these two reports did not reveal the clear structural criteria of lampbrush chromosomes found in animal cells.
In course of further studies, structures with a lampbrush-chromosome-like morphology have been described in the nucleoplasm of primary (giant) nuclei of the green algae Acetabularia mediterranea.
The occurrence of lampbrush type chromosomes was confirmed by light and electron microscopy in sections of algal cell fixed in situ and in spread preparations of isolated nuclear components. These chromosomes reveal typical loops up to 20µm long, chromomere-like nodules of 1-2µm in diameter and 2-4µm large axial globules.
Association of some of these chromosomes with nucleolar structures and with the nuclear envelope are also recognised. The light microscopically identified loops are correlated with distinct fibrillogranular structure observed in the sections and with the very long matrix units seen in the spread preparations.
8. Models of Chromosome Structure:
The chromosome of eukaryotic organism is basically made of two major components such as protein and nucleic acid like DNA. So chromosome is a nucleoprotein complex. But how the DNA protein complex builds up the chromosome structure is not clearly understood.
So initially it was under speculation and several models have been proposed time to time to explain the association of proteins with DNA. After then various studies and experiments have been done on chromosome structure to understand its biological architecture and again a new lots of model have been proposed. But all models are not universally accepted.
The various chromosome models may be grouped under three heads:
a. Multi-stranded model;
b. Single-stranded model;
c. Nucleosome-Solenoid model.
(a) Multi-Stranded Model:
In the multiple strand model, the chromosome is supposed to be made of several nucleoprotein strands. Cytologists on the basis of their observation have proposed a number of multi-stranded models. According to some, each chromosome consists of two chromatids which is divisible into two half chromatids.
Each half chromatid is again composed of two quarter chromatids. Each quarter chromatid is composed of four chromatin fibres.
Again each chromatin fibre is made of two strands. Each strand consists of a single DNA molecule plus the associated histone and non-histone proteins. DNA and proteins are held together by divalent cations like Ca++ and Mg++.
Thus one chromatid is made of 4x2x2x2 = 32 DNA molecule. Hence a chromosome with two chromatids is composed of 32 x 2 = 64 DNA molecules. According to this model the chromosome consists of 64 double helices of DNA arranged in a parallel manner and twisted together like the strands of rope (Fig. 13.16).
According to Ris histone is associated with DNA in some regular but unspecific fashion to form a DNA-histone or nucleoprotein fibril. Two nucleoprotein fibrils make up the elementary chromosome fibril. Two elementary chromosome fibrils wind spirally with each other to form a fibril.
Two fibrils constitute the chromonemata which forms several loops called chromomere. Each chromatid is made of four chromonemata. Hence each chromatid apparently has 16 elementary fibrils. Most of the evidence now indicate that chromosomes are not multi-stranded except giant polytene chromosome.
(b) Single-Stranded Model:
According to this model, chromosomes are single-stranded. Taylor (1957) proposed a single-stranded model according to which the chromosome is made of a long protein back bone from which DNA coils branch-off like the legs of a centipede.
Hence this model is known as Taylor’s centipede model (Fig. 13.17). The protein backbone is composed of two parallel layers of proteins and these layers can be pulled apart during replication.
It is thought that each layer has one strand of DNA helix on separation. On such a separated chromatid, a new chromatid could then be formed. The greatest demerit of this model is that it ignores the fact that genes are arranged in a linear fashion along the entire length of the chromosome. It is also inconsistent with genetic recombination data.
A second model was proposed jointly by Taylor and Freese. According to this model there are two protein spines instead of one. The DNA chains stretch between them like a Zigzag stair. In effect the DNA molecules are kept in position by the protein linkers (Fig. 13.14).
If the linkers become closely put together they would form the axis of chromosome and the DNA would be in the form of lateral loops. The only merit of this model that it can satisfy the concept that the genes are arranged in linear fashion.
Ris (1967) postulated a modified single- stranded model. According to this model DNA double helix binds with histone protein to form nucleoprotein fibrils. Folding of this fibrils takes place because of Ca++ bridge to form basic fibrils. The basic fibrils undergoes still further folding to form the chromosome (Fig. 13.18).
Du Praw, on the basis of his studies on human leucocytes under electron microscope, proposed a ‘Folded-Fibre Model’—to describe the structure of chromosome. According to this model chromosomes are made of chromatin fibres. Each chromatin fibre contains only one DNA double helix which is spirally coiled and coated with histone and non-histone proteins.
The fibre then becomes folded back longitudinally and transversely and thus intertwined and forms the body of a chromatid. Two sister chromatids remain held at the centromere. The folded fibre model is applies to both the interphase and the metaphase chromosomes.
During interphase folding is less and it is more at metaphase. This model is widely accepted and has been proved by various cytochemical, auto-radiographic and electron microscopic observation.
(c) The Nucleosome Model:
The nucleosome model is proposed by Roger Kornberg (1974). This model clearly explains the relationship of DNA and protein (Histone) as present within the chromosome. According to this model histones form core particle and DNA molecules coils around them so that the nucleoprotein fibre has beads on a string appearance.
(i) Histones:
There are five major types of histone molecules in the eukaryotic chromosome. These have been classified as histones H1,H2A,H2B,H3 and H4. The histones Eire the basic proteins of low Mr (mobility rate on electrophoresis) and account for just about the same mass as the DNA.
Histones are readily isolated by salt or acid extraction of chromatin. Each histone molecule consists of a hydrophobic core region with one or two basic arms.
Histone Hi is a very lysine rich protein of about 215 amino acids. Histones H2A and H2B are highly conserved and are known as the slightly lysine-rich histones. The most conserved of all are the arginine rich histones H3 and H4.
A special type of histone known as histone H5 is found in the nucleated erythrocyte of fish, amphibians and birds. It bears many similarities to histone Hi and is thought to maintain the highly repressed state of the chromatin in these non-dividing cell. In non-dividing cells of mammals histones H1° and H1e are present whereas histones H1a and H1b are present in large amounts only in dividing cells.
Histones may be methylated, phosphorylated, acetylated or ADP-ribosylated and some of these modifications of histone may take place by altering the charge on the molecule which may affect the interactions of histones with each other or with DNA.
For example there are six subtypes of histone H1 (H1a – e and H1°) giving rise to 14 different phosphorylated forms. Acetylation of histone H4 in particular causes unfolding of the nucleosome core his tones and is associated with transcriptionally active segment of chromatin.
About 20% of H2a histone is covalently linked with ubiquitin, a 76 residue polypeptide and forms a branched- chain protein known as UH2A which possibly control the gene expression. In sperm cell histones are replaced by other small basic proteins known as protamine’s.
(ii) Non-histone proteins:
Besides histones, some non-histone proteins are present in chromatin in an amount approximately equal to the histone. About 100 types of different non-histone proteins have been isolated from the chromatin. Some of these are the enzymes involved in replication and transcription or to form part of the nuclear envelope.
Other non-histone proteins can be classified into two categories like low mobility group (LMG) and high mobility group (HMG) of protein on electrophoresis.
Non-histone proteins are also basic protein like histone and they are present in multiple copies in the chromatin, i.e., they play a structural role. These proteins are not tightly associated with chromatin. The N-terminal and C-terminal part of non-histone proteins are separated by a short region which is rich in serine, glycine and proline. The most characterised are HMG1, HMG2, HMG 14 and HMG17.
(d) Experimental Evidence in Favour of Nucleosome Structure:
Several experimental studies have been made to prove the existence of nucleosome in chromatin structure.
The studies are:
(i) X-ray Diffraction Pattern Studies:
X-ray diffraction pattern of chromatin indicated the presence of a structure repeating every 10 nm.
(ii) Electron Microscopic Studies:
i. Electron microscopy of ruptured nuclei showed the presence of a series of spherical particles connected by a fine filament—the so called beads on a string. The beads have a diameter of 7-10 nm but the length of the filaments is variable.
Much work on the structure of sets nucleosome has been carried out with the virus SV40 (Simian Virus 40). DNA of Simian Virus 40 is a circular double- stranded molecule. When added to normal culture of cells, the DNA of SV40 may become integrated into the genome of the host. Normally viral DNA is devoid of nucleosomes. But under integrated condition viral DNA may form nucleosomal organisation.
Electron micrographs of SV40 infected cells also indicated the presence of nucleosome on viral DNA. The nucleosomal form of viral chromosome is known as minichromosome. Normally, the length of naked SV40 DNA is 1590 nm and that of the minichromosome is about 250 nm, it is clear that there has been six to seven fold packing of the DNA into the mini-chromosome.
(iii) Digestion of Chromatin with Micro-Coccal Nuclease:
It is already stated that the nucleosomes grossly appear as beads on string. It is obvious that beads are connected by non-beaded string or linker DNA which holds the nucleosomes. When a small fragments of DNA containing 4-5 nucleosomes are treated with micro-coccal nuclease, it gradually digests the linker DNA but nucleosome remains partially resistant to nuclease action.
An analysis of the size of the DNA showed that the spacing between successive nucleosomes was about 200 bp. On further digestion the size of the mono-nucleosome with about 200 bp DNA is reduced first to 166 bp and finally to 146 bp and H1is lost.
(iv) Crosslinking Studies:
Cross linking studies using di-methyl-suberimi-date have shown that in chromatin an octamer of histone composed of two molecules of H2A, H2B, H3 and H4 are present. Further studies have shown that one octamer is present per 200 bp DNA.
(v) Chromatin Digestion with Nuclease:
DNAse I (Nuclease) treatment makes nicks all along the length of DNA in chromatin. The nicks occur at ten base intervals. It means that the DNA is wrapped around a core of histones at a regular interval. Further studies involving the analysis of stoichemistry and X-ray crystallography have shown that one octamer is present per 200 bp DNA, i.e., per nucleosome. Each nucleosome is shallow, v-shaped structure around which a 146 bp core of DNA is wrapped making about one and three quarter turn.
(e) Nucleosome Structure:
All eukaryotic chromatin consists of nucleosomes. When interphase nuclei are ruptured by dipping them in a solution of low ionic strength, the chromatin fibres spill out of lysed nuclei. When isolated chromatin fibre is examined by electron microscope, it is seen that the chromatin fibre consists of a series of compactly organised ellipsoidal bead like particles.
The particles are joined by thin threads, a duplex of DNA. Actually a continuous duplex thread of DNA runs through the series of particles. The diameter of each particle is 110 A and the height is 60A. The beads or chromatin sub-unit is called nucleosome or Nu body.
Individual nucleosome (Fig. 13.19) consists of a 146 nucleotide pair length of core DNA. Core DNA wraps round core histone by one and three quarter turns (1 3/4).
Each core histone is composed of octamer (Fig. 13.20) containing two copies of the four histones H2A, H2B, H3 and H4. Histone H1 is present at one copy per nucleosome sealing the DNA entry/exit points to form a chromatosome of 166 bp and the remaining DNA forms the linker joining nucleosomes together to form oligonucleosomes.
The length of the linker varies from species to species and even within tissue. Linkers as short as 8 nucleotide pairs and as long as 114 nucleotide pairs have been reported.
A chromatin fibre is, therefore, made of a linear array of repeated nucleosome units plus a linker between every two nucleosomes (Fig. 13.21). Such a structural organisation constitutes a Poly-nucleosome. Under biological conditions, the nucleosome appear to be stable in position and to have little tendency to move along a length of DNA.
The nucleosome play a significant role in gene expression. Gene expression is related to the transcription which involves the unwinding of DNA and may require the fibre to unfold in restricted regions of chromatin that constitute a particular gene.
The linker DNA has no problem to unwind but the unwinding may be prevented where nucleosomes are present. It, therefore, seems inevitable that transcription of active gene must involve a structural change to unwind DNA.
Again during transcription, the enzyme, RNA polymerase is essential to move along the length of template. On the basis of observation it is clear that an important structural change occurs when a gene is intensely transcribed.
In case of the rRNA genes the nucleosomes are entirely displaced. Hence it seems that RNA polymerases displaces the nucleosome at the point of transcription but that the histone octamer immediately recaptures its position unless another RNA polymerase is present to prevent it from doing so.
During replication the DNA is free of nucleosomes. Once DNA has been replicated, nucleosomes are quickly generated on both the duplicates.
The diameter of a double helix of DNA is 2 nm whereas the diameter of metaphase chromatid is much thicker. Hence it is obvious that DNA undergoes a higher order of supercoiling (Fig. 13.22). The diameter of a nucleosome is about 10 nm. Therefore, in the first state of condensation, the nucleosomes are packed into a spiral or solenoid arrangement with six nucleosome per turn.
The pitch of the solenoid is 11 nm and the faces of the nucleosome are approximately parallel to the solenoid axis. The fifth histone H1, i.e., is bound to the DNA on the inside of the solenoid. The solenoid structure then forms a number of loops around a central core or scaffold or a matrix which is made of an ill-defined fibrous protein network.
The scaffold proteins also include two abundant proteins of Mr (Mobility rate) 1,70,000 and 1,35,000. The larger is DNA topoisomerase II and the smaller binds MARs (Matrix attachment regions) in co-operative fashion.
Both initiation and continued replication of DNA occur in association with matrix proteins and topoisomerase II binding sites Eire found on matrix associated DNA. The binding sites for topoisomerase II are called Scaffold Associated Regions (SARs).
9. Chemical Structure of Chromosomes:
Chemical analysis of eukaryotic chrosomes has shown that they are composed of deoxyribonucleic acid (DNA), ribonucleic acid (RNA) histone and non-histone proteins and certain metallic ions like Ca++, Mg++, etc. Primarily, chromosome contains about 90% DNA—basic protein forming a nucleoprotein complex and 10%.
RNA non-histone protein, although it is variable according to the metabolic state of nucleus. Nucleoprotein complex constitutes the backbone of chromosome while RNA non-histone protein complex is sometimes regarded as residual chromosome. The ratio of DNA— basic, protein in chromatin—is nearly 1 : 1 and remains constant over a wide range of plants and animals.
The histones axe joined with phosphate of DNA as salt linkage. The protamine’s are bound to the DNA by ionic bonds. Besides this Mg++,Ca++ ions are supposed to maintain the chemical architecture of chromosome intact.
10. Biological Importance of Chromosome:
The chromosomes are considered the very important biological organisation because of the following reasons:
i. The genetic material DNA is localised in the chromosome and its contents are relatively constant from one generation to the next.
ii. The chromosomes retain their structure, individuality and continuity throughout the life-cycle of organism.
iii. The chromosomes maintain and replicate the genetic information contained in their DNA molecule and this information is transcribed at the right times in proper sequence into the specific types of RNA molecules which directs the synthesis of different types of proteins to form a body- form like the parents.
iv. The chromosomes form the only link between two generations and plays a significant role in the development of an organism from the zygote.