This article provides information about the Fine or Ultrastructure of Chromosome!
Contents
[I] Chromosome is the multistranded body:
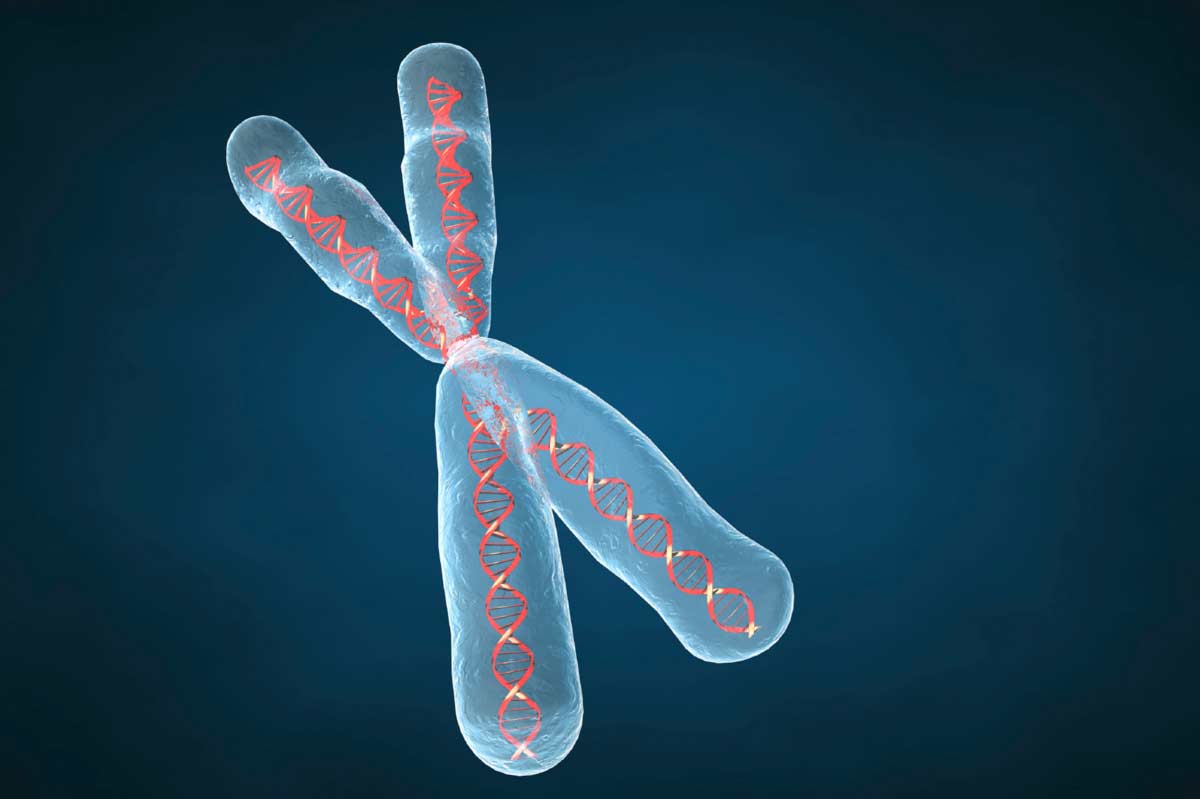
The smallest unit of chromosome (by electron microscopy) is the fibril, which is 100 A thick.
This fibril contains two DNA double helixes, separated by a space about 25 A across, and the associated protein.
The next largest unit of the chromosome is the quarter chromatid.
The quarter chromatid consists of four 100 A fibrils, so that it is about 400 A thick having eight double helixes of DNA and the associated protein. Two quarter chromatids form a half chromatid, which therefore contains 16 double helixes of DNA.
A chromatid contains two half chromatids, bringing the total number of helixes in a chromatid to 32 and its diameter is about 1600 A before DNA synthesis or replication. After replication the chromosome contains 64 double helixes of ‘DNA with an approximate diameter of 3200 A. The thickness is quite variable from cell to cell and from organism to organism due to differences in coiling of the chromosomes.
The DNA molecule consists of one old helix and one new helix. The chromosome of a higher organism may contain eight or more double helixes, depending upon its degree of subdivision.
[II] Single continuous fibre of nucleoprotein (Folded-fibre organization):
A chromosome consists of a single long chain of DNA and protein, upto several hundred microns long. For example, the total length of DNA— protein making up 46 chromosomes of man has been estimated as 50 cm.
The fibre is extremely coiled. The lowest level is represented by 100 A superhelix. This helical fibre is then folded many times and irregularly entwined to form the body of a chromatid, and there is no subchromatid-strand structure. The packing of the fibres may depend upon Ca++ and Mg++ ions.
Thus the basic unit of chromosome is a DNA-protein superhelix having a diameter of about 100 A and these units make up 250 A fibres as seen in electron microscope. In addition, presence of histone protein and divalent cations (Ca++ and Mg++) is essential for coiling of such fibres.
[III] Folded fibre model of chromosome:
‘Folded-fibre model’ of chromosome structure was proposed by Du Praw (1966), in which the chromosomal unit is regarded as a single DNA-protein fibre, which is repeatedly folded back on itself both longitudinally and transversely to make up the body of the chromatid. The model defines a ‘unit chromatid’ as a chromatin fibre of variable length, of folding configuration and of replication mechanism.
It corresponds to one continuous Watson-Crick DNA molecule. The single DNA helix of the unit chromatid is thought to be wrapped with polypeptides in P-configurations (e.g., lysine-rich histones) and then super coiled to make up the 200 to 300 A chromatin fibres.
Presumably super coiling is induced and maintained by cross-linking with a-helical proteins, such as arginine rich histones. During interphase S period, the unit chromatid replicates sequentially at two or more replication forks, a process that involves local unwinding of the tertiary and secondary helices.
Each pair of daughter fibres corresponds to a pair of metaphase chromatids, which are held together by unreplicated portions of the fibre, especially in the centromere region (middle figure 6.10).
As proposed in the ‘folded-fibre model,’ the protein components of the unit chromatids are regarded having not only histones but also various DNA-linked nonhistone proteins, including such enzymes as RNA polymerase, NAD synthetase, nucleoside triphosphatase, phospho proteins, histone acetylases, and possibly DNA polymerase and enzymes of nuclear oxidative phosphorylation.
These molecules may be arranged on DNA filaments in very orderly functional arrays. Presumably histones are able to account for DNA super coiling. DNA is itself responsible for linear continuity of a chromosome. It is the DNA-linked proteins which mediate chromosome condensation.
[IV] Synaptinemal complex (SC):
In certain organisms like crayfish, locust, pigeon and rat and in early meiotic prophase of spermatogenesis shows a linear complex of three parallel strands of DNA in the central axis of the chromosome. These strands form synaptinemal complex.
A single synaptinemal complex is a ribbon-like object, consisting of two parallel dense rods (each of 450 A diameter) separated uniformly at a distance of 1000 to 1200 A, and connected to a median longitudinal element by transverse fibres. This structure was on first discovered in males of crayfish, pigeons, cats and human beings but not present in male Drosophila.
This complex appears to be associated with the synapsis of the chromosomes during meiotic prophase. The synaptinemal complex is interposed between pairing homologues and can be considered the structural basis of pairing. Lateral elements of SC start to form at leptonema, while median element appears with pairing at zygonema. During pairing, two homologues remain separated by a 0.15 to 0.2 µm space that is occupied by SC.