In this article we will discuss about the Physicochemical Organization of Chromosome.
The major chemical components of the chromosomes are DNA, proteins (histones and non-histones) and small amount of RNA. Besides these, calcium and some other minerals are also present. Histones are basic proteins that are charged positively at neutral pH. Non-histone proteins are acidic. Histones are small proteins containing some 100-200 aminoacids, about 20 to 30 per cent of which are lysine and arginine.
The positive charges on histones enable them to bind to DNA by electrostatic attraction to the negatively charged phosphate groups in the sugar-phosphate backbone of DNA. Histones also bind tightly to each other. Histones in both DNA-histones and histone to histone bindings play important role in chromatin structure-DNA packaging.
They are present in chromatin of all higher eukaryotes in the amount equal to that of DNA weight/weight. The histones present in the nuclei of all higher eukaryotes including man are of five different types. These are called H1, H2a, H2b, H3 and H4. In the sperms of some eukaryotes, however, a special class of small basic proteins called protamines replaces histones.
There is a remarkable constancy of histones H1, H2a, H2b, H3 and H4 in all types of organisms and among widely divergent species. This shows that histones have been highly conserved during evolution. They are present in molar ratios of approximately 1H1, : 2H2a : 2H2b : 2H3 : 2H4.
They are specifically complexed with DNA to form basic structural unit of chromatin-the nucleosome. Acidic proteins or non-histone proteins consist of several heterogeneous proteins of different compositions and are thought to be involved in the regulation or expression of some genes or groups of genes.
The quantitative measurements of DNA have been made in a number of cases. The most convenient unit of measurement of DNA is picogram (10-12) which is equivalent to 31 cms of double helical DNA.
The quantity of DNA varies greatly in cells of different kinds of organisms, as for example, human diploid cell will have 174 cm of DNA (5-6 picograms), Trillium cells will have 37 metres (120 picograms) and Drosophila salivary gland chromosome will have 90 metres per cell (293 picograms).
It is one thing to determine the chemical composition of the chromosome, but quite another to work out the molecular arrangement of nucleic acids in the chromosomes. Chromosome thickness is usually hundred times that of DNA and length of DNA found in a chromosome is several hundred times that of chromosome.
How does this condensation takes place? What are the components that are involved in packaging process? What is the packaging scheme? These are the most challenging and frustrating problems for the biologists today.
For any acceptable model of chromosome structure, there are two basic requirements:
(i) The linear sequence of genes must be preserved, and
(ii) The replication and segregation of DNA should be explained.
As regards the physicochemical organization of chromosomes, several models have been proposed which may be discussed under the following groups:
(i) Multiple strand or multineme models, and
(ii) Single strand or unineme models.
Multiple Strand Models:
The multiple strand models have been proposed in light of the observed variation in the thickness of the nucleoprotein fibrils, from 30 A to 200 A or even 500 Å. In multineme model, the chromosome is supposed to contain many DNA-protein fibrils.
The evidence in favour of this model comes from the studies on the chromosomes of Tradescantia (8 fibrils of 200 Å each/chromatid), mosquito (16 strands per chromatid). Amoeba proteus (8 strands/chromatid).
Ris Model:
From the electron microscopic studies of the chromosomes of plants and animals during meiosis and mitosis, Ris (1957) concludes that chromosome is multistranded (polytene) and it is built like a cable with numerous identical sub-units. Kaufmann and Mcdonald (1957) supported Ris.
However, the possibilities that the chromosome is formed of single long DNA cannot be ruled out by these observations because multiple strands observed in electron micrograph might represent a single strand folded forth and back upon itself many times.
Single Strand Models:
In bacteria and viruses there is no well organised chromosome. The genetic material in these cases appears to be DNA. DNA molecule in bacteria forms bacterial chromosome and is sometimes termed as chromoneme or genophore. In bacteriophage, DNA molecule may be 50 µ long. In bacterium it may be 1 to 1.5 mm long and in large chromosomes the DNA molecules may be several centimetres long.
At present, most investigators agree that chromosomes have a single DNA-histones system. Whitehouse and Hastings (1965) suggest that a chromatid can be looked upon as one DNA molecule. Taylor working with the chromosome structure of vicia root tips and Gall and Callan (1962) working with the lamp-brush chromosomes of triturus oocytes suggest that chromatid is a single stranded structure.
They are of the view that many genes may be represented in one DNA molecule. There appears no evidence at present to support the occurrence of more than one DNA molecule in normal chromosome. If it is supposed that single molecule of DNA is extended along the entire length of chromosome the DNA duplication seems to be difficult in view of long distance which it should unwind and fold during the process of division.
Taylor-Freese Model:
J.H. Taylor (1957) suggested that chromosome consists of a long two layered protein backbone from which DNA coils branch off just like the legs of a centipede. Taylor (1959) proposed a new model based on the one suggested by Freese (1958).
In the model, several linkers have been shown, some of which could open at certain operational sites for the control and order of the nucleotide sequences (Figs. 9.14 and 9.15). Several polynucleotide chains may be involved to stabilize the structure during duplication. The linkers join the terminal phosphates of polynucleotide chains.
If it is supposed that a chromosome has a single DNA molecule then the question arises as to how the histones fit into chromosome structure. The unequal spacing of two polynucleotide strands of DNA leaves two spiral grooves or furrows.
Watson and Crick in 1953 have suggested that the molecular structure of DNA was such that fully extended polypeptide chains (histone strands) could be fit in the spiral grooves along the helix of the polynucleotide chains. Feughelman et al (1955) confirmed this. Wilkins (1957) also tentatively suggested that in DNA-histone complex there might be a polypeptide strand in each groove of DNA.
Proteins (histones) of chromosomes probably play the following roles:
(i) They provide characteristic morphological shapes and structures to chromosomes.
(ii) Histones of chromosomes show species specificity and their main function consists in protecting the DNA strands from breakage and allowing them to fold into condensed configurations.
(iii) They also play a part presumably in meiosis at the time of chromosome pairing. Moses and Coleman (1964) have found that pairing cores of the homologous chromosomes, when seen under electron microscope, at first meiosis are composed of proteins and it may be that the close pairing of homologous chromosomes comes about through some sort of interaction between histones and proteins of pairing cores.
(iv) They also play important role in the control of gene action and in cell differentiation.
Folded Fibre Model of Du Praw:
According to “folded fibre model” of Du Praw, each eukaryotic chromosome is composed of a single greatly elongated and highly folded nucleoprotein fibril of 100 Å thickness (Fig. 9.16). This nucleoprotein fibril is composed of a single, linear, double stranded DNA molecule of 20 Å thickness which remains wrapped in equal amounts of histones and non-histones and variable amounts of different kinds of RNA.
The nucleoprotein fibril is coiled in packing ratio of 6 : 1 to form a short fibre of 80-100 A diameter (packing ratio is the ratio of fully extended length to packed length). This fibre is called Type A Fibre. Type A fibre, in turn, is supercoiled in packing ratio of 10 : 1 to form a Type B fibre of 200—350 Å diameter which is similar to bead seen in electron micrographs of chromatin fibres.
Du Praw has tried to explain how the nucleoprotein fibril of chromosome replicates during cell division and how it is held at the centromere where the DNA segment remains un-replicated until the anaphase (Fig. 9.17).
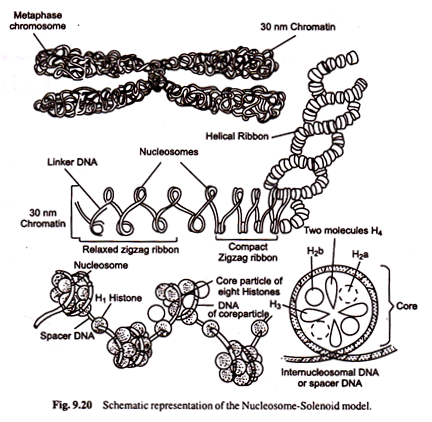
Nucleosome-Solenoid Model:
The chromosome during prophase appears as a delicate fibre consisting of one long molecule of DNA complexed with proteins. In fact, in chromatin the DNA is in tightly packed state representing at least a hundred fold contraction. This is achieved by several levels of folding.
When observed under electron microscope, the chromatin appears to be made up of a series of ellipsoidal beads joined by thread-like structures (Fig. 9.18). These beads are nucleosomes or nu- bodies joined by DNA, called linker DNA. Nucleosomes were first described by A. Olins, D. Olins and by C.L.F. Woodcock in 1970s.
When chromatin is digested with endonuclease, the enzyme, first attacks linker DNA in the regions between nucleosomes. As a result of this, the nucleosomes are released but the DNA in the nucleosome is protected from digestion by endonuclease.
The pieces of DNA associated with nucleosomes are about 200 bps long. This indicates that there are about 200 bps in DNA from linker to linker. Further digestion by endonucleases causes digestion of linker DNA-leaving behind nucleosome cores with a piece of DNA that is about 146 base pairs long. Nucleosome core is disc shaped or flat sphere with a diameter of about 110 Å and thickness of about 50 Å.
The nucleosome core contains a histone octamer of two molecules each of histones H2a, H2b, H3 and H4 and DNA invariably forming 1 and 3/4 (1 3/4) coils around the core. The association of core DNA with histone octamer is tight one and the DNA is well protected against enzyme action (i.e., endonuclease enzyme does not digest any DNA associated with histones).
The size of linker DNA varies in different species. H1 histone is located outside the nucleosome cores. It holds the two strands of DNA together as they enter and leave core. It is bound to the edges of the nucleaosome discs across the linker DNA and stabilizes about 166 base pairs long DNA. H1 histone molecules seal off two turns of DNA around the histone octamer core (Fig. 9.19).
The most complex level of packaging involves higher-order coiling of the DNA-histone complexes to produce chromosome morphology as shown in the figure. The supercoiled nucleosome fibre is called solenoid. A number of acidic non-histone chromosomal proteins are thought to be involved in further coiling and packing of solenoid.
The exact roles of non-histone proteins in chromosome structure and function are unknown. Some of these proteins are believed to have structural roles in the formation of 2,000 to 4,000 Å super-solenoid.
When viewed under electron microscope, the DNA- Protein complex in chromosomes appears as a fibre or nucleoprotein filament of about 10 nm diameter (Fig. 9.20). The solenoid structure has a diameter of about 300 Å. Obviously the DNA can be compacted extensively by superimposing coils on coiled structures during cell division.
The chromatin exists in a more highly coiled state than the 100 Å thick nucleofilament while the packing of DNA in the nucleosomes is less well understood in the higher order of folding of the nucleosome in the chromosomes. Above the level of nuclesome and 100 Å nucleoprotein filament, there is a next level of packing which forms 300 A thick chromatin fibre.
Examination of isolated 300 Å chromatin fibre reveals the complex nature of the chromosome structure.
The arrangement of nucleosome in the fibre is difficult to define. The next level of packing beyond 300 Å chromatin fibres are poorly understood. The diameter of interphase chromosome may be 3,000Å while the diameter of a metaphase chromosome may be 7,000 Å.
To achieve these levels of compaction, the simplest concept is that 300 Å fibre becomes looped and coiled like a rope in various ways to give morphologies characteristic of these chromosome.