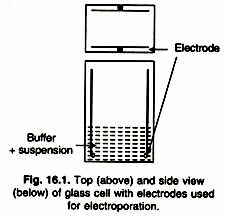
Some of the methods used to repair inappropriate damaged bases are as follows:
(a) Direct Reversal of Base Damage (b) Excision Repair.
(a) Direct Reversal of Base Damage:
Spontaneous addition of a methyl group (CH3-) (an example of alkylation) to Cs followed by domination to a T is the most frequent cause of point mutations in humans.
Fortunately, most of these changes are repaired by enzymes which are known as glycosylases that remove the mismatched T restoring the correct C. The DNA backbone need not be broken for this.
DNA used to get damaged by alkylation in cancer chemotherapy (“chemo”) due to some of the drugs used also damage DNA by alkylation. Some of the methyl groups can be removed by a protein encoded by our MGMT gene. The removal of each methyl group requires another molecule of protein as the protein can only do it once.
Each of the myriad types of chemical alterations to bases requires its own mechanism to correct. The cell needs are more general mechanisms capable of correcting all sorts of chemical damage with a limited toolbox. The mechanisms of excision repair this requirement.
(b) Excision Repair:
In this process the damaged base or bases are removed and then replaced with the correct ones in a localized burst of DNA synthesis. There are three modes of excision repair, each of which employs specialized sets of enzymes namely. Base Excision Repair (BER), Nucleotide Excision Repair (NER), and Mismatch Repair (MMR). Figure 4.4 gives base excision repair (BER) steps.
The steps and by players of BER are:
(i) Removal of the damaged base by a DNA glycosylase.
(ii) Removal of its deoxyribose phosphate in the backbone which produces a gap.
(iii) Replacement with the correct nucleotide. This relies on DNA polymerase beta, one of at least 11 DNA polymerases encoded by our genes.
(iv) Ligation of the break in the strand. Two enzymes are known that can do this; both require ATP to provide the needed energy.
Nucleotide Excision Repair (NER) differs from BER in several ways. It uses different enzymes. NER removes a large “patch” around the damage. Figure 4.5 explains nucleotide excision repair (NER) steps.
The steps and key players of NER are:
(i) The damage one or more protein factors recognize. These assemble at the location.
(ii) The DNA is unwound which produces a “bubble”. The enzyme system that does this is Transcription Factor IIH, TFIIH, (which also functions in normal transcription).
(iii) Cuts are made on both the 3′ side and the 5′ side of the damaged area so the tract containing the damage can be removed.
(iv) A fresh burst of DNA synthesis – using the intact (opposite) strand as a template – fills in the correct nucleotides. The DNA polymerases responsible are designated polymerase delta and epsilon.
(v) A DNA ligase covalently inserts the fresh piece into the backbone.
Xeroderma Pigmentosum (XP) is a rare inherited disease of humans. It predisposes the patient to pigmented lesions on areas of the skin exposed to the sun and an elevated incidence of skin cancer. XP can be caused by mutations in any one of several genes.
Nucleotide-excision repair (NER) proceeds most rapidly in cells whose genes are being actively transcribed and on the DNA strand that is serving as the template for transcription.
XPB, XPD, and several other gene products are involved in this enhancement of NER. The genes for two of them are designated CSA and CSB. The CSB product associates in the nucleus with RNA polymerase II, the enzyme responsible for synthesizing messenger RNA (mRNA), are providing a molecular link between transcription and repair.
In E. coli, proteins UvrA, UvrB, and UvrC are involved in removing the damaged nucleotides. The gap is then filled by DNA polymerase I and DNA ligase. In yeast, the proteins similar to Uvr’s are named RADxx such as RAD3, RADIO, etc. (“RAD” stands for “radiation”).
Mismatch Repair (MMR) deals with correcting mismatches of the normal bases. Figure 4.6 gives mismatch repair (MMR) steps.
It can enlist the aid of enzymes involved in both base-excision repair (BER) and nucleotide-excision repair (NER) as well as using enzymes specialized for this function. Recognition of a mismatch requires several different proteins including one encoded by MSH2. Cutting the mismatch out also requires several proteins, including one encoded by MLH1.
The process of repairing starts with the protein MutS which binds to mismatched base pairs. MutL is recruited to the complex and activates MutH which binds to GATC sequences. Activation of MutH cleaves the unmethylated strand at the GATC site. Then, the segment from the cleavage site to the mismatch is removed by exonuclease (with assistance from helices II and SSB proteins). If the cleavage occurs on the 3′ side of the mismatch, this step is carried out by exonuclease I.
It degrades a single strand only in the 3′ to 5′ direction. If the cleavage occurs on the 5′ side of the mismatch, exonuclease VII or RecJ is used to degrade the single stranded DNA. Mismatch repair is very expensive and inefficient as the distance between the GATC site and the mismatch could be as long as 1,000 base pairs.
Homologs of MutS and MutL have been found in yeast, mammals, and other eukaryotes. MSH1 to MSH5 are homologous to MutS; MLH1, PMS1 and PMS2 are homologous to MutL. Colon cancer relate to mutations of MSH2, PMS1 and PMS2.