Biotechnology deals with techniques of using live micro-organisms, plant or animal cells or their components or enzymes from organisms to produce products and processes (services) useful to human beings.
The term biotechnology was coined in 1917 by a Hungarian Engineer, Karl Ereky to describe a process for large scale production of pigs.
Gene manipulation is a fast emerging science. It started with the development of recombinant DNA molecules. It is named variously as DNA manipulation biotechnology, recombinant DNA technology and genetic engineering.
The technology mostly involves cutting and pasting of desired DNA fragments. It is based on two important discoveries in bacteria:
(i) Presence of plasmids in bacteria which can undergo replication along with and independent of chromosomal DNA.
(ii) Restriction endonucleases (Arber, Nathan and Smith 1970; Nobel Prize in 1978) which can break DNA at specific sites.
They are appropriately called molecular scissors. Berg (1972) was able to introduce a gene of SV-40 into a bacterium with the help of lambda phage. Berg is often considered “father of genetic engineering”. He was awarded Nobel Prize in 1980. The science of recombinant technology took birth when Cohen and Boyer (1973) were able to introduce a piece of gene containing foreign DNA into plasmid of Escherichia coli.
Old Biotechnology (Traditional Biotechnology):
Microorganisms were first used to produce some organic compounds like citric acid. They were also used to produce antibodies. The levels of production of penicillin yield has been improved But the types of products are not changed. They remain the same as those obtained from the natural strains/cell lines. In all these processes, only the natural capabilities of the organisms and cells are exploited. These activities are called old biotechnology.
Modern Biotechnology:
Human insulin is also produced from a transgenic Escherichia coli stain that contains and expresses the insulin gene. Proteins produced by transgenes are called recombinant proteins. The production technologies based on genetic engineering are termed as modern biotechnology. It developed during 1970.
Definition of Biotechnology:
A definition of biotechnology which covers both traditional views and modern molecular biotechnology has been given by European Federation of Biotechnology (EFB). According to EFB. “Biotechnology is the integrated use of biochemistry, microbiology and engineering sciences in order to achieve technological (industrial) application of the capabilities of microorganisms, cultured tissues/cells and parts thereof’. Thus definition of biotechnology involves two common factors. First the use of biological agents and second the product or service is generated for the well being of humans.`
I. Principles of Biotechnology:
Two Main Techniques of Modern Biotechnology:
The two main techniques that gave birth to modem biotechnology are as follows:
(a) Genetic Engineering:
It includes techniques to alter the nature of genetic material (DNA and RNA) to introduce these into host organisms and thus change the phenotype of the host organism.
(b) Chemical Engineering:
It involves maintenance of sterile microbial contamination free condition in chemical engineering processes to have growth of only the desired microorganism/eukaryotic cell in large quantities for the manufacture of biotechnological products such as antibiotics, vaccines, enzymes, medicines, hormones, etc.
1. Conceptual Development of the Principles of Genetic Engineering:
Genetic engineering is a kind of biotechnology which deals with the manipulation of genetic material by man in vitro.
Two Main Discoveries:
Genetic engineering is based on two important discoveries in bacteria.
(i) Presence of plasmids in bacteria which can undergo replication along with and independent of chromosomal DNA.
(ii) Restriction endonucleases (Arber, Nathan and Smith 1970; Nobel Prize in 1978) which can break DNA at specific sites. They are appropriately called molecular scissors or biological scissors.
Role of Paul Berg:
In 1972 genetic engineering was started by Paul Berg. Berg (1972) was able to introduce a gene of SV-40 virus into a bacterium with the help of lambda phage. Berg is often considered “father of genetic engineering”. He was awarded Nobel Prize in 1980.
The technique of genetic engineering includes:
(i) Formation of ‘recombinant DNA’ (rDNA).
(ii) Use of gene cloning.
(iii) Gene transfer. It permits to isolate and introduce only one or a set of desirable genes without introducing undesirable genes into the target organism.
A piece of DNA which is introduced into the alien (foreign) organism would not be able to multiply itself in the organism but when it gets incorporated into the genetic material of the recipient, it may multiply and be inherited along with the host DNA, because the alien piece of DNA has become part of chromosome which possesses the ability to replicate.
There is a specific DNA sequence called the origin of replication in a chromosome that is responsible for initiating replication. Thus, an alien DNA linked with the origin of replication can replicate and multiply itself in the host organism. It is also called the cloning, i.e., forming multiple identical copies of any template DNA.
2. Construction of the First Artificial Recombinant DNA Molecule:
The first recombinant DNA was constructed by Stanley Cohen and Herbert Boyer in 1972. They cut the piece of DNA from a plasmid carrying antibiotic resistance gene in the bacterium Salmonella typhimurium. Cutting of a piece of DNA from a plasmid was with the help of restriction enzymes (also called molecular scissors or chemical scalpels).
The piece of foreign DNA cut from the plasmid was linked with the plasmid DNA acting as vector. Linking of the piece of foreign DNA with vector was done with the help of the enzyme DNA ligase which acts on cut DNA molecules and join their ends. This newly formed DNA having integrated fragment of antibiotic resistant gene is called recombinant DNA.
The vectors is used to transfer, recombinant DNA to E. coli. This transfer of recombinant DNA is similar to the transfer of malaria parasite from diseased person into the healthy person through female Anopheles mosquito (acts as an insect vector).
When this recombinant DNA is transferred into Escherichia coli, it could replicate in the new host cell in the presence of DNA polymerase enzyme and make multiple copies of recombinant DNA. The ability to multiply copies of antibiotic resistance gene in E. coli was termed as cloning of antibiotic resistance gene in E. coli.
Three basic steps in creating genetically modified organism (GMO) or transgenic organism. GMO contains a foreign gene/segment of DNA. These three basic steps are as follows.
(i) Identification of DNA with desirable genes.
(ii) Introduction of the identified DNA into the host.
(iii) Maintenance of introduced DNA in the host and transfer of the DNA to its progeny.
II. Tools of Recombinant DNA Technology:
Three types of “biological tools” are used in the formation of recombinant DNA. These are as follows:
(A) Enzymes
(B) Cloning Vectors (Vehicle DNA)
(C) Competent host (for transformation with recombinant DNA)
(A) Enzymes:
(1) Lysing Enzymes:
These enzymes are used to open up the cells to get DNA for genetic experiments. Lysozyme is usually used to dissolve the bacterial cell wall. In plant cell, cell wall is made up of cellulose, while in fungi, it is made up of chitinase.
(2) Cleaving Enzymes:
These enzymes are used to break DNA molecules. They are of three types— exo-nucleases, endonucleases and restriction endonucleases.
(a) Exo-nucleases:
They remove nucleotides from the terminal ends (either 5′ or 3′) of DNA in one strand of duplex.
(b) Endonucleases:
They make cuts at specific position within the DNA. These enzymes do not cleave the ends and involve only one strand of the DNA duplex.
(C) Restriction endonucleases:
They cut DNA duplex at specific points. Their single stranded free ends are called ‘sticky ends’ (Fig. 11.3) which can be joined end to end by DNA ligases.
Restriction Endonucleases — The Molecular Scissors:
Restriction endonuclease was isolated for the first time by W. Arber in 1962 in bacteria. They are called “molecular scissors or biological scissors”. In 1978 Arber, Smith and Nathan were awarded the Nobel Prize for the discovery of restriction endonuclease.
They recognize the base sequence at palindrome sites in DNA duplex and cut its strands. For example, restriction endonuclease EcoR 1 found in the colon bacteria E. coli, recognizes the base sequence GAATTC in DNA duplex and cuts its strands between G and A as shown below :  Only restriction enzymes type II are used in gene manipulation for two reasons,
Only restriction enzymes type II are used in gene manipulation for two reasons,
(a) No ATP is needed for the cleaving action,
(b) It makes cleavage or cut in both the strands of DNA molecule.
(1) Types of Restriction Endonucleases:
Three main types of restriction endonucleases are type I, type II and type III.
Type I Restriction Endonucleases:
These enzymes consist of 3 different subunits. They require ATP, Mg2+ and S-adenosyl methionine for restriction. Type I restriction endonucleases recognize specific sites within the DNA but do not cut these sites. Thus they are not used in recombinant DNA technology.
Type II Restriction Endonucleases:
These enzymes are simple and require Mg2+ ions for restriction. Out of the three types, only type II restriction enzymes are used in recombinant DNA technology.
Тyре III Restriction Endonucleases:
These enzymes consist of two different subunits. They require ATP, Mg2+ and S-adenosyl methionine for restriction. They recognize specific sites within DNA but do not cut these sites, therefore, these restriction endonucleases are not used in recombinant DNA technology. They have intermediate properties between type I and type II.
Differences Between Тyре I, Тyре II and Type III Restriction Endonucleases:
| Type I | Type II | Тyре III | |
| 1. | Enzyme structure consists of 3 different subunits. | Enzyme structure is simple. | Enzyme structure consits of two different sub units. |
| 2. | They require ATP, Mg2+ S-adenosyl-methionine for restriction. | They require Mg2+ ions for restriction. | They require ATR Mg2+ and S-adenosyl, methionine for restriction. |
| 3. | They recognize specific sites within DNA but do not cut these sites. | They recognize specific sites within the DNA and cut these sites. | They recognize specific sites within the DNA but do not cut these sites. |
| 4. | They are not used in recombinant DNA technology. | They are used in recombinant DNA technology. | They are not used in recombinant DNA technology. |
(2) Nomenclature of Restriction Enzymes:
(i) Type II restriction enzymes are named for the bacterium from which they have been isolated,
(ii) The first letter used for the enzyme is the first letter of the bacterium’s genus name (in italics),
(iii) Then comes the first two letters of its species (also in italics),
(iv) The fourth letter of the name of enzyme is first letter of the strain. It is written in capital,
(v) The end of the name indicates the order in which the enzyme was isolated. It is written in Roman number. For example, the enzyme Eco R1 was isolated from the bacterium Escherichia coli RY13. Enzyme Eco R1 is named as follows.
The capital letter E comes from the genus Escherichia. The letters со. are from the species coli. The letter R is from RY13 (strain). The Roman number 1 indicates that it was the first enzyme isolated from the bacterium E. coli RY13. The discovery of restriction endonuclease enzymes led to Nobel Prize for W. Arber, H. Smith and D. Nathan in 1978.
Some examples of type II enzymes are given in the table 11.1 showing names of restriction enzymes (endonucleases), source (organism from where they have been isolated), their recognition sequence and site of cleavage. 
(3) Functioning of Restriction Endonuclease:
The foundations of recombinant DNA (rDNA) were laid by the discovery of restriction enzymes. These enzymes are present in many bacteria where they function as a part of their defence mechanism called the Restriction Modification System. Molecular basis of this system was explained first by Wemer Arber in 1965.
The Restriction-Modification system consists of two components;
(i) A restriction enzyme which identifies the introduced foreign DNA and cuts into pieces called restriction endonucleases. The term ‘restriction’ refers to the function of these enzymes in restricting the propagation of foreign DNA of bacteriophages (viruses that attack bacteria) in the host bacterium,
(ii) The second component is a modification enzyme that adds a methyl group to one or two bases usually ‘within’ the sequence reorganized by the restriction enzyme.
Once a base in a DNA sequence is modified by addition of a methyl group, the restriction enzymes fail to recognize and could not cut that DNA. This is how a bacterium modifies and therefore, protects its own chromosomal DNA from cleavage by these restriction enzymes.
The first restriction endonuclease was Hin d II (hin-dee-two). Its functioning depended on a specific DNA nucleotide sequence. It was isolated from Haemophilus influenza Rd. It was found that Hin d II always cut DNA molecules at a particular point by recognising a specific sequence of six base pairs.
This specific base sequence is known as the recognition sequence for Hin d II. It produces blunt ends. Besides Hin d II, today we know more than 900 restriction enzymes that have been isolated from over 230 strains of bacteria each of which recognises different recognition sequences.
(4) Palindromic Nucleotide Sequence:
The restriction endonuclease inspects the length of a DNA sequence. Once it recognises specific sequence, it binds to the DNA and cuts each of the two strands of the double helix at specific points in their sugar phosphate back bones. Special sequence in the DNA recognised by restriction endonuclease is called palindromic nucleotide sequence.
Restriction endonuclease recognizes palindromic sequences in DNA and cuts them. The palindromes are groups of letters that form the same words when read in both directions forward and backward.
Examples:
The palindromes in DNA are base pair sequences that are the same when read forward (left to right) or backward (right to left) from a central axis of symmetry. For example, the following sequences read the same on the two strands in 5’—> 3′ direction. This is also true when we read in the 3’—> 5′ direction.
Restriction enzymes cut the strand of DNA a little away from the centre of the palindrome sites but between the same two bases of the opposite strands. This leaves single stranded unpaired bases at cut ends. These ends with unpaired bases are called sticky ends or cohesive ends (Fig 11.3). The latter are named so because they form hydrogen bonds with their complementary cut counter parts. The sticky ends facilitate the action of the enzyme DNA ligase.
Gel Electrophoresis (Separation and Isolation of DNA Fragments):
After the cutting of DNA by restriction enzymes, fragments of DNA are formed. Separation of DNA fragments according to their size or length is done by a technique called gel electrophoresis developed by A. Tiselius in 1937. Electrophoresis is a technique of separation of molecules such as DNA, RNA or protein on the basis of their size, under the influence of an electrical field, so that they migrate in the direction of electrode bearing the opposite charge, viz., positively charged molecules move towards cathode (-ve electrode) and negatively charged molecules travel towards anode (+ve electrode) through a medium/matrix.
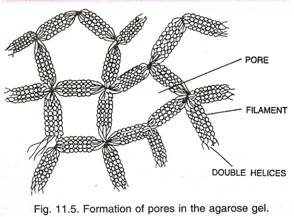
Electrophoresis performed in a gel matrix, so that molecules of similar electric charge can be separated on the basis of size, is called gel electrophoresis. Now a days the most commonly used matrix is agarose which is a polysaccharide extracted from sea weeds. DNA fragments separate according to size through the pores of agarose gel. Hence the smaller the fragment size the farther it moves. Agarose dissolves in hot water when this solution is cooled, double helices form and become arranged laterally and produce thick filaments. These filaments become cross- linked to form the gel (Fig. 11.5). Pore size depends on agarose concentration.
The separated DNA fragments can be seen only after staining the DNA with a compound known as ethidium bromide (EtBr) followed by exposure to UV radiation as bright orange coloured bands. The separated bands of DNA are cut out from the agarose gel and extracted from the gel piece. This process is called as elution (removal of adsorbent). These purified DNA fragments are used in constructing recombinant DNA by linking them with cloning vectors.
(B) Cloning Vectors (Vehicle DNA or carrier of DNA):
The vectors are DNA molecules that can carry a foreign DNA segment and replicate inside the host cell. Vectors may be plasmids, a bacteriophages (viruses that attack bacteria), cosmids, Yeast artificial chromosomes (YACs), Bacterial artificial chromosomes (BACs) and viruses. There are also some shuttle vectors. Out of these vectors, the most commonly used cloning vectors are plasmids and viruses.
(1) Plasmid Vectors:
Plasmids were discovered by Willium Hays and Joshua Lederberg (1952). These are extra-chromosomal, self-replicating, usually circular, double-stranded DNA molecules, found naturally in many bacteria and also in some yeast. Although plasmids are usually not essential for normal cell growth and division, they often confer some traits on the host organism, for example, resistance to certain antibiotics or toxins that can be a selective advantage under certain conditions.
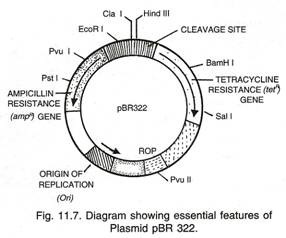
The plasmid molecules may be present as 1 or 2 copies or in multiple copies (500-700) inside the host organism. These naturally occurring plasmids have been modified to serve as vectors in the laboratory. The most widely used, versatile, easily manipulated vector pBR 322 is an ideal plasmid vector.
pBR322 Vector (Fig. 11.7):
This was the first artificial cloning vector constructed in 1977 by Boliver and Rodriguez. It is widely used in gene cloning experiments.
Nomenclature:
In pBR 322 plasmid
p — denotes that it is a plasmid;
BR — stands for Boliver and Rodriguez who constructed this plasmid;
322 — is a number given to distinguish this plasmid from others developed in the same laboratory. For example, there are plasmids pBR 325, pBR 327, pBR 328, pBR 345 etc.
Features that is required to facilitate cloning into a Vector:
(i) Origin of Replication (Ori):
Origin of replication (Ori) is a specific sequence of DNA bases which is responsible for initiating replication. A prokaryotic DNA has a single origin of replication while eukaryotic DNA may have more than one origin of replication.
(ii) Selectable Markers:
Some genes called “selectable markers” help in selecting those host cells which contain the vectors (trans-formants) and eliminating the non-trans formants. Transformation is a process through which a piece of DNA is introduced in a host bacterium. Generally, the genes encoding resistance to antibiotics such as tetracycline, ampicillin, kanamycin or chloramphenicol etc. are useful selectable markers for E. coli. The common E. coli cells are not resistant against any of these antibiotics. Plasmid pBR 322 has two resistance genes — ampicillin resistance (ampR) and tetracyclin resistance (tetR ) which are considered useful for selectable markers.
(iii) Cloning Sites (Recognition Sites):
Plasmid pBR 322 has a variety of unique recognition sites for restriction endonucleases. Two unique sites, Pst I and Pvu I are located within the ampR gene and Bam HI, Sal I, etc. are within tetR gene (Fig 11.7). Some other unique restriction sites are Eco RI, Cla I, Hind III, Pvu II, rop codes for the proteins involved in the replication of the plasmid.
The presence of restriction sites within the markers tetR and аmрR permits an easy selection for cells transformed with the recombinant pBR322. Insertion of the DNA fragment into the plasmid using enzyme Pst I or Pvu I places the DNA insert within the gene ampR; this makes аmрR non-functional. Bacterial cells containing such a recombinant pBR322 will be unable to grow in the presence of ampicillin, but will grow on tetracycline. Similarly, when restriction enzyme Bam HI or Sal I is used, the DNA insert is placed within the gene tetr making it non-functional. Bacterial cells possessing such a recombinant pBR322 will, therefore, grow on ampicillin but not on tetracycline.
Due to inactivation of antibiotics, selection of recombinants becomes burdensome process because it requires simultaneous plating on two plates having different antibiotics. Thus, alternative selectable marker is developed to differentiate recombinants and non-recombinants on the basis of their ability to produce colour in the presence of a chromogenic substance. Now a recombinant DNA is inserted in the coding sequence of an enzyme β- galactosidase.
This casues inactivation of the enzyme which is called insertional inactivation. If the plasmid in the bacterium does not have an insert, the presence of a chromogenic substrate gives blue coloured colonies. Presence of insert results into insertional inactivation of the p-galactosidase and, therefore, the colonies do not produce any colour, these colonies are marked as recombinant colonies.
(iv) Vectors for Cloning Genes in Plants and Animals:
A soil-inhabiting plant bacterium, Agrobacteriun tumefaciens, a pathogen (disease causing agent) of several dicot plants is able to transfer a piece of DNA known as ‘T-DNA’. The T-DNA causes tumours. The tumours are called crown galls. Tumour formation is induced by Ti plasmid (Ti for tumour inducing).
As gene transfer occurs without human effort, the bacterium is called natural genetic engineer of plants. Similarly retroviruses (cause leukosis or sarcoma types of cancer) in animals including humans are able to change normal cells into cancerous cells.
The tumour including Ti plasmid of Agrobacterium tumefaciens have been modified into cloning vector which is not pathogenic to the plants, however, it is still able to use the procedure to deliver genes of our interest into various plants. Similarly retroviruses are used to carry desirable genes into animal cells. Thus once a gene or DNA fragment is joined to a suitable vector it is transferred into a bacterial plant or animal host where it undergoes multiplication.
(2) Bacteriophage Vectors:
Bacteriophages (are simply phages) are viruses that attack bacterial cells by injecting their DNA into these cells. The injected DNA is selectively replicated and expressed in the host bacterial cell resulting in a number of phages which burst out of the cell (lytic pathway) and reinfect neighbouring cells.
This ability to transfer DNA from the phage genome to specific bacterial hosts during the process of bacterial infection gave scientists the idea that specially designed phage vectors would be useful tools for gene cloning experiments. Several bacteriophages are being used as cloning vectors but most commonly used are lambda (X) phage and M13 phage.
(a) Lambda Phage Vector:
It has a double-stranded, linear DNA genome of 48, 502bp, in which the 12 bases at each end are unpaired but complementary. These ends are, therefore, sticky or cohesive and are referred to as the cos sites (cohesive end sites). These sites are important for packaging DNA into phage heads. The lambda genome remains linear in the phage head, but within E. coli cells the two cohesive ends join to form a circular molecule necessary for replication. These vectors allow cloning of dna fragments upto 23 Kb length (= Kilobase— is a unit designating the length of a nucleic acid sequence, e.g., 1 Kb = 1000 nucleotide long base sequence).
(b) M13 Phage Vector:
It is filamentous phage which infects E. coli having F-pili. Its genome is a single stranded, circular DNA of 6407 bp. Foreign DNA can be inserted into it without disrupting any of the essential genes. After the Ml 3 phage DNA enters the bacterial cell, it is converted to a double-stranded molecule known as the replicative form or RF, which replicates until there are 100 copies in the cell and single-stranded copies of the genome are produced and extruded from the cell as M13 particles.
The major advantages of developing vectors based on M13 are that its genome is less than 10Kb length. The RF can be purified and manipulated exactly like a plasmid. In addition, genes cloned in M13 based vectors can be obtained in the form of single-stranded DNA.
Why are bacteriophage vectors more advantageous than plasmid vectors?
(i) Bacteriophage vectors can be used for large DNA fragments and
(ii) These can easily be detected at the time of cloning experiments.
(C) Competent Host (For Transformation with Recombinant DNA):
(a) DNA Mediated Gene Transfer (Vector Mediated Gene Transfer):
Competent host is essential for transformation with recombinant DNA (Fig. 11.12). Transformation “a process by which a cell takes up naked DNA fragment from the environment, incorporates it into its own chromosomal DNA and finally expresses the trait controlled by the incoming DNA”. The tools described earlier in this chapter will result in the generation of recombinant DNA (rDNA) molecules in the laboratory. Finally, the propagation of these DNA molecules must occur inside a living system or a host.
Many kinds of host cells, including E. coli, yeast, animal and plant cells, are available for genetic engineering and the kind of host cell to be used mainly depends on the aim of the cloning experiment. For the expression of some eukaryotic proteins, eukaryotic cells may be the preferred hosts. Yeasts have been used extensively for functional expression of eukaryotic genes because they offer several advantages. Yeasts are the simplest eukaryotic organisms and like bacteria are single-celled, genetically well-characterized, easy to grow and manipulate.
They can be grown readily in both small culture vessels and large scale bioreactors. Plant and animal cells may also be used as hosts in gene manipulation experiments and for protein expression either in tissue culture or as cells in the whole organism to create genetically modified plants and animals. Since DNA is a hydrophilic molecule, it cannot pass through membranes, so the bacterial cells must be made capable to take up DNA.
This is done by treating them with a specific concentration of a divalent cation, such as Calcium which increases the efficiency with which DNA enters the bacterium through pores in its cell wall. Recombinant DNA (rDNA) can then be forced into such cells by incubating the cells with recombinant DNA on ice, followed by placing them briefly at 42°C (heat shock), and then putting them back on ice. This enables the bacteria to take up the recombinant DNA.
Transfer of DNA into eukaryotic cell is called transfection.
(b) Direct or Vector less Gene Transfer:
It is the process of gene transfer into the host cell without using a vector. This is possible by the following four important methods.
1. Micro-injection:
In this method foreign DNA is directly injected into the nucleus of animal cell or plant cell by using micro needles or micro pipettes. It is used in oocytes, eggs and embryo. Alec Jeffreys (1993) of Human Genome Centre, Michigan University U.S.A has cured mice that inherited a neuromuscular disease which is like muscular dystrophy of humans.
2. Electroporation:
Electroporation is the formation of temporary pores in the plasma membrane of host cells by using lysozyme or calcium chloride. These pores are used for introduction of foreign DNA.
3. Chemical Mediated Gene Transfer:
In this method certain chemicals such as polyethylene glycol (PEG) help foreign DNA to enter the host cell.
4. Biolistic Method or Gene gun Method:
Biolistic is a means of introducing DNA into cells that involves bombardment of cells with high-velocity micro projectiles coated with DNA. In biolistic method tungsten or gold particles, coated with foreign DNA are born barded into target cells at a very high velocity. Although this method is suitable for plants yet this technique is also used to insert genes into animal that promote tissue repair into cells (particularly cancer of mouth) near wounds.
This method failed to make an impression in treatment of genetic disorder but made great impact in the field of vaccine development.
After being introduced briefly to the tools in recombinant DNA let us describe the processes to create recombinant DNA.