Contents
Molecular Markers:
A molecular marker is a DNA sequence in the genome which can be located and identified. As a result of genetic alterations (mutations, insertions, deletions), the base composition at a particular location of the genome may be different in different plants.
These differences, collectively called as polymorphisms can be mapped and identified. Plant breeders always prefer to detect the gene as the molecular marker, although this is not always possible. The alternative is to have markers which are closely associated with genes and inherited together.
The molecular markers are highly reliable and advantageous in plant breeding programmes:
i. Molecular markers provide a true representations of the genetic makeup at the DNA level.
ii. They are consistent and not affected by environmental factors.
iii. Molecular markers can be detected much before development of plants occur.
iv. A large number of markers can be generated as per the needs.
Basic principle of molecular marker detection:
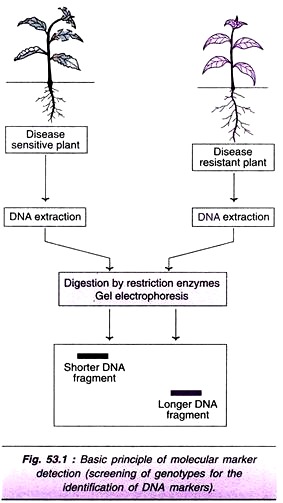
Let us assume that there are two plants of the same species—one with disease sensitivity and the other with disease resistance. If there is DNA marker that can identify these two alleles, then the genome can be extracted, digested by restriction enzymes, and separated by gel electrophoresis. The DNA fragments can be detected by their separation.For instance, the disease resistant plant may have a shorter DNA fragment while the disease — sensitive plant may have a longer DNA fragment (Fig. 53.1).
Molecular markers are of two types:
1. Based on nucleic acid (DNA) hybridization (non-PCR based approaches).
2. Based on PCR amplification (PCR-based approaches).
Markers Based On DNA Hybridization:
The DNA piece can be cloned, and allowed to hybridize with the genomic DNA which can be detected. Marker-based DNA hybridization is widely used. The major limitation of this approach is that it requires large quantities of DNA and the use of radioactivity (labeled probes).
Restriction fragment length polymorphism (RFLP):
RFLP was the very first technology employed for the detection of polymorphism, based on the DNA sequence differences. RFLP is mainly based on the altered restriction enzyme sites, as a result of mutations and re-combinations of genomic DNA. An outline of the RFLP analysis is given in Fig. 53.2, and schematically depicted in Fig. 53.3.The procedure basically involves the isolation of genomic DNA, its digestion by restriction enzymes, separation by electrophoresis, and finally hybridization by incubating with cloned and labeled probes (Fig. 53.2).
Based on the presence of restriction sites, DNA fragments of different lengths can be generated by using different restriction enzymes. In the Fig. 53.3, two DNA molecules from two plants (A and B) are shown. In plant A, a mutations has occurred leading to the loss of restriction site that can be digested by EcoRI.
The result is that when the DNA molecules are digested by the enzyme Hindlll, there is no difference in the DNA fragments separated. However, with the enzyme EcoRI, plant A DNA molecules is not digested while plant B DNA molecule is digested. This results in a polymorphic pattern of separation.
Markers Based on PCR Amplification:
Polymerase chain reaction (PCR) is a novel technique for the amplification of selected regions of DNA .The advantage with PCR is that even a minute quantity of DNA can be amplified. Thus, PCR-based molecular markers require only a small quantity of DNA to start with.
PCR-based markers may be divided into two types:
1. Locus non-specific markers e.g. random amplified polymorphic DNA (RAPD); amplified fragment length polymorphism (AFLP).
2. Locus specific markers e.g. simple sequence repeats (SSR); single nucleotide polymorphism (SNP).
Random amplified polymorphic DNA (RAPD) markers:
RAPD is a molecular marker based on PCR amplification. An outline of RAPD is depicted in Fig. 53.4. The DNA isolated from the genome is denatured the template molecules are annealed with primers, and amplified by PCR.
Single short oligonucleotide primers (usually a 10-base primer) can be arbitrarily selected and used for the amplification DNA segments of the genome (which may be in distributed throughout the genome). The amplified products are separated on electrophoresis and identified.
Based on the nucleotide alterations in the genome, the polymorphisms of amplified DNA sequences differ which can be identified as bends on gel electrophoresis. Genomic DNA from two different plants often results in different amplification patterns i.e. RAPDs. This is based on the fact that a particular fragment of DNA may be generated from one individual, and not from others. This represents polymorphism and can be used as a molecular marker of a particular species.
Amplified fragment length polymorphism (AFLP):
AFLP is a novel technique involving a combination of RFLP and RAPD. AFLP is based on the principle of generation of DNA fragments using restriction enzymes and oligonucleotide adaptors (or linkers), and their amplification by PCR. Thus, this technique combines the usefulness of restriction digestion and PCR.
The DNA of the genome is extracted. It is subjected to restriction digestion by two enzymes (a rare cutter e.g. Msel; a frequent cutter e.g. EcoRI). The cut ends on both sides are then ligated to known sequences of oligonucleotides (Fig. 53.5).
PCR is now performed for the pre-selection of a fragment of DNA which has a single specific nucleotide. By this approach of pre-selective amplification, the pool of fragments can be reduced from the original mixture. In the second round of amplification by PCR, three nucleotide sequences are amplified.
This further reduces the pool of DNA fragments to a manageable level (< 100). Autoradiography can be performed for the detection of DNA fragments. Use of radiolabeled primers and fluorescently labeled fragments quickens AFLP.
AFLP analysis is tedious and requires the involvement of skilled technical personnel. Hence some people are not in favour of this technique. In recent years, commercial kits are made available for AFLP analysis. AFLP is very sensitive and reproducible. It does not require prior knowledge of sequence information. By AFLP, a large number of polymorphic bands can be produced and detected.
Sequence tagged sites (STS):
Sequence tagged sites represent unique simple copy segments of genomes, whose DNA sequences are known, and which can be amplified by using PCR. STS markers are based on the polymorphism of simple nucleotide repeats e.g. (GA)n, (GT)n, (CAA)n etc. on the genome. STS have been recently developed in plants. When the STS loci contain simple sequence length polymorphisms (SSLPs), they are highly valuable as molecular markers. STS loci have been analysed and studied in a number of plant species.
Microsatellites:
Microsatellites are the tandemly repeated multi-copies of mono-, di-, tri- and tetra nucleotide motifs. In some instances, the flanking sequence of the repeat sequences may be unique. Primers can be designed for such flanking sequences to detect the sequence tagged microsatellites (STMS). This can be done by PCR.
Sequence characterized amplified regions (SCARs):
SCARs are the modified forms of STS markers. They are developed by PCR primer that are made for the ends of RAPD fragment. The STS-converted RAPD markers are sometimes referred to as SCARs. SCARs are useful for the rapid development of STS markers.
Molecular Marker Assisted Selection:
Selection of the desired traits and improvement of crops has been a part of the conventional breeding programmes. This is predominantly based on the identification of phenotypes. It is now an accepted fact that the phenotypes do not necessarily represent the genotypes. Many a times the environment may mark the genotype. Thus, the plant’s genetic potential is not truly reflected in the phenotypic expression for various reasons.
The molecular marker assisted selection is based on the identification of DNA markers that link/ represent the plant traits. These traits include resistance to pathogens and insects, tolerance to abiotic stresses, and various other qualitative and quantitative traits. The advantage with a molecular marker is that a plant breeder can select a suitable marker for the desired trait which can be detected well in advance. Accordingly, breeding programmes can be planned.
The following are the major requirements for the molecular marked selection in plant breeding:
i. The marker should be closely linked with the desired trait.
ii. The marker screening methods must be efficient, reproducible and easy to carry out.
iii. The analysis should be economical.
Molecular Breeding:
With rapid progress in molecular biology and genetic engineering, there is now a possibility of improving the crop plants with respect to yield and quality. The term molecular breeding is frequently used to represent the breeding methods that are coupled with genetic engineering techniques.
Improved agriculture to meet the food demands of the world is a high priority area. For several years, the conventional plant breeding programmes (although time consuming) have certainly helped to improve grain yield and cereal production.
The development of dwarf and semi-dwarf varieties of rice and wheat have been responsible for the ‘Green Revolution’, which has helped to feed millions of poverty-stricken people around the world. Many developments on the agriculture front are expected in the coming years as a result of molecular breeding.
Linkage analysis:
Linkage analysis basically deals with studies to correlate the link between the molecular marker and a desired trait. This is an important aspect of molecular breeding programmes. Linkage analysis has to be carried out among the populations of several generations to establish the appropriate linkage. In the earlier years, linkage analysis was carried out by use of isoenzymes and the associated polymorphisms. Molecular markers are now being used. The techniques employed for this purpose have already been described.
Quantitative Trait Loci:
These are many characteristics controlled by several genes in a complex manner. Some good examples are growth habit, yield, adaptability to environment, and disease resistance. These are referred to as quantitative traits. The locations on the chromosomes for these genes are regarded as quantitative trait loci (QTL).
The major problem, the plant breeder faces is how to improve the a complex character controlled by many genes. It is not an easy job to manipulate multiple genes in genetic engineering. Therefore, it is a very difficult and time consuming process. For instance, development of Golden Rice (with enriched pro-vitamin A) involving the insertion of just three genes took about seven years.
Arid and Semi-Arid Plant Biotechnology:
The terms arid zone is used to refer to harsh environmental conditions with extreme heat and cold. The fields have limited water and minerals. It is different task to grow plants and achieve good crop yield in arid zones. Semi-arid regions are characterized by unpredictable weather, inconsistent rainfall, long dry seasons, and poor nutrients in the soil.
Most parts of India and many other developing countries (Africa, Latin America, and Southeast Asia) have semi- arid regions. Crops like sorghum, millet, groundnut and cowpea are mostly grown in semi-arid tropics. Besides unpredictable weather, biotic and abiotic stresses contribute to crop loss in these areas.
The biotechnological approaches for the breeding programmes in the semi-arid regions should cover the following areas:
i. Development of crops that are tolerant to drought and salinity.
ii. Improvements to withstand various biotic and abiotic stresses.
iii. Micro-propagation techniques to spread economically important plants which can withstand harsh environmental conditions.
Some success has been achieved in improving sorghum, millet and legume crops that are grown in semi-arid regions. Genetic transformation in sorghum was possible by using micro projectile method.
Greenhouse and Green-home Technology:
Greenhouse literally means a building made up of glass to grow plants. Green houses are required to grow regenerated plants for further propagation and for growing plants to maturity. Greenhouses are the intermediary stages involving the transitional step between the plant cultures and plant fields. The purpose of greenhouses is to acclimatize and test the plants before they are released into the natural environment.
The plants are grown in greenhouse to develop adequate root systems and leaves so as to withstand the field environment. The greenhouses are normally equipped with cooling systems to control temperature. Greenhouses have chambers fitted with artificial lights. It is possible to subject the plants to different lighting profiles. In recent years many improvements have been made in the development of more suitable greenhouses. These include the parameters such as soil, and humidity.
The major limitation of greenhouse technology is an increase in CO2 production that in turn increases temperature. Some approaches are available to control temperature. Green home technology is a recent development. In this case, temperature is controlled by using minimum energy.