Here is a list of seven viruses that are found in plants: 1. Tobacco Mosaic Virus (TMV) 2. Potato Virus X (PVX) 3. Potato Virus Y (PVY) 4. Cucumber Mosaic Virus (CMV) 5. Cauliflower Mosaic Virus (CaMV) 6. Potato Leaf Roll Virus (PLRV) 7. Mosaic Disease of Sugarcane.
Contents
1. Tobacco Mosaic Virus (TMV):
TMV is the most serious pathogen causing mosaic on tobacco leaves. It is transmitted by artificial inoculation but not by insect vectors. TMV is the most resistant virus known so far of which the thermal death point is 90°C for 10 minutes.
TMV is very stable and the preparations of unpreserved plant juice have been found to retain infectivity even after 50 years. But a temperature- sensitive, nitrous acid-induced mutant is much less stable.
They are very stable to heat. Some strains can retain infectivity after 10 minute exposures to more than 90°C. Dilutions of 106 of expressed tobacco sap can be infectious. Purified virus preparations may be preserved at 4°C for long periods by mixing a few drops of chloroform that inhibits the growth of microorganisms.
Symptoms of TMV:
TMV damages the solanaceous plants. However, it can infect the other plants too. Several strains of TMV has also been reported. After infection, it develops symptoms of lightening of leaf colour along the veins in early stages. Thereafter, it turns into light and dark green mosaic symptoms.
Along the veins green colour turns into dark green and the internal region turns into chlorotic. Sometimes dark green blisters appear in the leaf blade. If the plants are infected early in season they become stunted. However, symptoms vary with varieties of tobacco. The virus reduces the yield as well as quality of the products i.e. the nicotine content is decreased by 20-30 %.
Virus Structure of TMV:
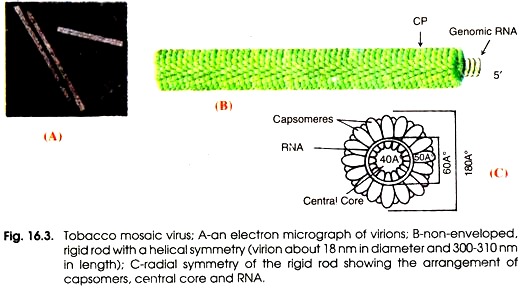
Fig. 16.3 shows an electron micrograph of TMV viron (A), a non- enveloped rigid rod with a helical symmetry (B) and a radical symmetry of the rigid rod showing the arrangement of capsomers, central core and RNA (C). Franklin (1957) have described the structure of TMV. It is rod-shaped helical virus measuring about 280 × 150 µm with a molecular weight of 39 × 106 Daltons. The virion is made up of 2,130 protein subunits of identical size.
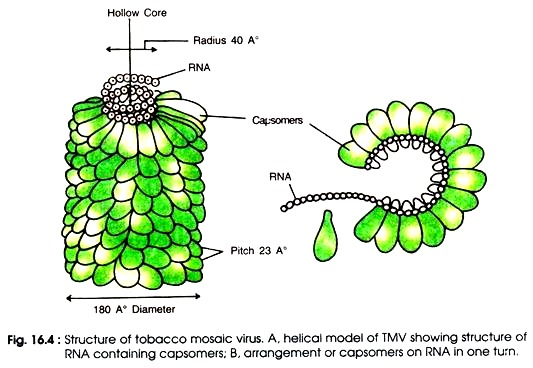
The protein subunits are arranged around a central hole of 4 nm (40Å) (Fig. 16.4 A-B). Each protein subunit is made up of a single polypeptide chain which possesses 158 amino acids, the molecular weight of which is 17,500 Daltons. Inside the protein capsid there is a single stranded RNA molecule which is also spirally coiled to form helix.
Virus RNA consists of 6,500 nucleotides. In one turn the RNA contains 49 nucleotides. Total number of protein subunits counting in three turns is 49 i.e. 49/3 subunits per turn. Therefore a single protein subunit is linked with 3 nucleotides of RNA. Arrangement of capsomers on RNA is shown in Fig.16.4 B.
Genome of TMV:
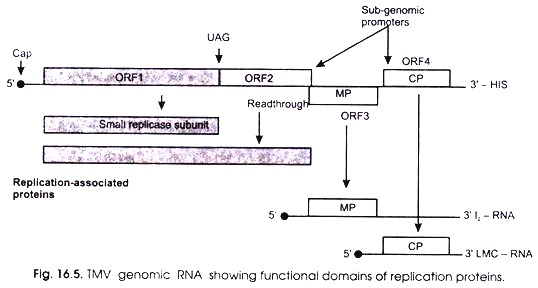
Fig. 16.5 shows the functional domains of replication proteins of TMV genomic and sub-genomic RNA. The genome of TMV is a mono-partite, linear, (+) sense mRNA of 6.3-6.5 kb that produces five proteins during virus infection. The nucleotide sequence and genome organization of Tobamovirus were determined over 20 years ago. But there is still controversy concerning the number of proteins encoded by the TMV genome.
The RNA possesses a cap (m7G5’pppG) at the 5′-end and a tRNA-like structure charged with the amino acid histidine (His) at the 3′-end. Two RNA polymerase proteins are produced after direct translation of the genomic RNA. The 5′ proximal open reading frame (ORF1) encodes a 126 kDa protein involved in replication, which contains motifs characteristic of methyl transferase (MET, capping enzyme) and helicase (HEL) domains.
Read-through of the amber terminator of ORI 2 yields a 183 kDa protein which is considered to be the viral polymerase. Only the 126 and 183 kDa proteins are required for replication of the genomic RNA or various defective RNAs generated in vitro.
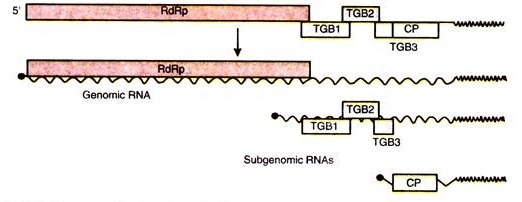
Another protein is produced due to ribosomes by-passing a leaky stop codon UAG. Besides, it also contains an RN A-dependent RNA polymerase (RdRp) domain. Two other proteins are also produced from the viral RNA; these are called movement protein (MP) from ORF3 and capsid protein (CP) from ORF4.
During the infection by TMV, three sub-genomic RNAs (sgRNAs) have been identified. All of which have been found in association with polyribosomes. The smallest of these sgRNAs, designated LMC-RNA (low molecular mass component), encodes the 3′ proximal 17-5 kDa capsid protein (CP) (0RF4).
The other largest sgRNA, designated I2 (intermediate-class RNA 2), encodes the 30 kDa viral movement protein (MP) (0RF3). The I1 sgRNA (intermediate-class RNA 1) contains an ORF encoding a 54 kDa protein, which coincides with the read-through portion of the 183 kDa protein. Unlike the other TMV-encoded proteins, the I1 sgRNA expressed 54 kDa protein has not been detected in vivo.
Sulzinski (1985) have characterized this third sub-genomic TMV RNA (i.e. 11-RNA. It is associated with polyribosomes; therefore, it is presumed that it acts as a messenger RNA in vivo. The 11-RNA was shown to be a subset of the TMV genome, representing the 3′-half of the molecule. The 11 -RNA was thus shown to be a distinct RNA species and not a class of heterogeneous molecules of approximately the same size.
The II-RNA 5′ terminus is residue 3405 in the genome. After an un-translated sequence of 90 bases, an AUG codon at residues 3495-3497 initiates a protein of 54 kDa terminating at residue 4915. Thus, the amino acid sequence of the 54 kDa protein is coincident with those residues of the carboxy terminus of the well-known 183 kDa TMV protein.
During the derivation of viral genome organizations from the sequences of their genomes, separate ORFs of less than 10 kDa are ignored, unless they occurred in several strains of the same virus or in different viral species in the same genus. An ORF which is referred to as ORF6 in the genomes of TMV and the tobamoviruses and Tobacco mild green mosaic virus have been described by Canto (2004) which encodes a protein of about 4-5 kDa.
Multiplication of TMV:
A virus needs to enter the vascular system of the plant for successful colonization of an entire plant. The process of cell-to-cell movement of TMV is relatively slow; it takes one too few hours to multiply in a cell and move to the next cell. Systemic transport of TMV normally occurs through the phloem sieve elements where viruses move passively with the flow of photosynthates.
After rapid systemic spread of the virus in the phloem, it moves from the phloem to surrounding cells where it reproduces and spreads by cell-to-cell movement. The time between initial infection of one or few cells and systemic infection of the plant varies from few days to few weeks depending on the virus, host plant, and environmental conditions.
Thereafter, TMV moves into the neighboring cells. Depending on the virus, its genomes or the virions are transported into neighboring cells through small channels called plasmodesmata that form connections between cells. Many viruses produce movement proteins that modify the plasmodesmata channels and facilitate viral movement into neighboring cells.
a. Gene expression:
The virion RNA of TMV is infectious and serves as both the genome and viral messenger RNA. The genome encodes at least four proteins. The methyltransferase/ helicase (mt/hel) and RNA-dependent RNA polymerase (RdRp) involved in vims replication are in the 5′ half of the genome.
The first one is directly translated from genomic RNA, and the second one is produced by read-through of the termination codon of the first gene. The other two, a movement protein and the capsid protein, are expressed from separate sub-genomic mRNAs.
b. Replication:
After penetration, TMV enters the host cells (Fig. 16.6A), and replicates in cytoplasm of the infected cells. Inside the cell, the protein coat dissociates and viral nucleic acid becomes free in the cell cytoplasm.
The sites for different steps of the viral multiplication and formation of new viruses have not yet been completely determined. But the studies suggest that after becoming free in the cell cytoplasm the viral-RNA moves into the nucleus (possibly into the nucleolus) (B).
The viral-RNA first induces the formation of specific enzymes called ‘RNA polymerases’, in the presence of which the single-stranded viral-RNA synthesizes an additional RNA strand called ‘replicative RNA’ (C).
This RNA strand is complementary to the viral genome and serves as ‘template’ for producing new RNA single strands of which are the copies of the parental viral-RNA (D). The new viral RNAs are released from the nucleus into the cytoplasm and serve as mRNAs (E). In cooperation with ribosomes and tRNA of the host cell, each mRNA directs the synthesis of protein sub-units (F).
After the production of desired protein sub-units (capsomeres) (G-H), the new viral RNA is considered to organize the protein subunits around it resulting in the formation of complete virions (I-K). Unlike virulent bacteriophages, no lysis of the host cell takes place. The host cells remain alive and viruses move from one cell to the other causing systemic infection.
Protein Synthesis of TMV:
Takeba (1975) demonstrated the direct entry of TMV into the isolated protoplast from mesophyll cells of tobacco. After making entry, RNA rapidly starts uncoating by removing the subunits from the capsid by using host enzymes. The parental RNA is localized in nucleus but not in cytoplasm.
It performs two important functions:
(i) It acts as mRNA and directs and the synthesis of protein, and
(ii) Functions as template for synthesis of complementary strand.
The virus RNA utilizes the amino acids, ribosomes and tRNA of the host and synthesizes the complementary strand and proteins i.e. coat proteins of 17,500 Daltons and two other polypeptides (of molecular weight 160,000 and 140,000 Daltons).
The ratio of nucleic acid and protein differs with each virus. Nucleic acid is about 5-40% of the virus and protein 60-70%. Each protein subunit of TMV consists of 158 amino acids making a total number to about 17,531.
Transmission of TMV:
TMV is transmitted through the cell sap of host and enters a new host through wound incision. Wound is caused in plant due to various cultural operations such as clipping or topping the shoot. It is not seed transmitted but acts as seed contaminant. It is also transmitted by wind and water. Various control methods of the disease are regular roguing diseased plants and weeds, sanitation and use of resistant varieties.
Control of TMV:
TMV occurs in tobacco- growing areas, and is considered to be one of the most important tobacco viruses economically. Insects are not important in its spread. Control is by crop rotation, effective sanitation and use of resistant cultivars of tobacco. For the first time, TMV has been used to demonstrate for development of coat protein-mediated resistance, replicase-mediated resistance, and movement protein-mediated resistance.
Demonstration of TMV RNA as Genetic Material:
Gierer and Schramm (1956) and Fraenkel-Conrat and Singer (1957) have demonstrated that RNA is the genetic material in TMV by using viral strains. They selected two strains of TMV e.g. TMV A and TMV B, separated protein and RNA and reconstructed new TMV particles as given in Fig. 16.7.
i. Type A particles contained RNA of TMV A + protein of TMV B
ii. Type B particles contained RNA of TMV B + protein of TMV A
These reconstructed particles were inoculated on healthy tobacco leaves to develop symptoms. Symptoms developed on both kinds of inoculated tobacco leaves according to types of viral RNAs, but not according to capsid proteins. This confirms that in TMV, RNA acts as genetic material, not the capsid protein.
2. Potato Virus X (PVX):
Potexviruses belong to the family Flexiviridae of the Group IV (+) single-stranded RNA as per classification of ICTV. PVX is distributed throughout the world. Development of symptoms on different varieties of potato differs.
There are several strains of PVX. The infected tubers transmit the viral particles. It can remain alive for about 5 months but can be inactivated at 74°C. A large number of solanaceous plants such as tobacco, datura, Solanum nigrum, egg plants, pepper, tomato, etc. can also be infected by PVX.
In India research on viruses and viral diseases was started in 1940 when import of seeds from Europe was banned and available seed stocks degenerated within the short period.
The Indian Council of Agricultural Research started a scheme for production of disease-free seed potato which came into operation at Kufri (now the Central Potato Research Institute) at Shimla in 1940. PVX was first isolated in 1945 by Vasudeva and Lal. Thereafter, several groups of this virus were reported.
Symptoms of PVX:
PVX is also called by the other names such as Solarium virus, potato latent virus mottle virus. The infected potato shows a wild mosaic between the veins on foliage. Sometimes it is not visible clearly. In many cells PVX do not impart any symptoms. The infected plants become dwarf and deformity in foliage occurs. Symptoms produced on different hosts differ considerably. In the varieties Epicure, crest, Key Edward the virus produces top necrosis.
Structure of PVX:
The virion is non-enveloped, flexuous, filamentous, 470-1000 nm or more long and 12-13 nm in diameter. Virions are associated with helper virus. But it does not depend on them during replication (Fig. 16.8). Capsid is made up of identical protein subunits forming a helix of 3.3 nm pitch with a hole of 3 nm diameter. Possibly, a single subunit is associated with 3 or 4 nucleotides.
Strains of PVX exist in nature which can be detected serologically or on the basis of symptoms in tobacco and Capsicum plants, thermal inactivation point (TIP 68-67°C), and reaction in potato cultivars determining top necrotic response.
Genome of PVX:
The virus codes for 5 ORF(s). The virion RNA is infectious and serves as both the genome and viral mRNA. It consists of a single linear genome of (+) sense ssRNA having 5.8- 9 kb of nucleotides. The 3′ terminus is polyadenylated.
In some genera the 5′ end is capped. They encode for 3 to 6 proteins. RNA-dependent RNA polymerase (RdRp) is translated directly from the genomic RNA. The other ORFS are transcribed presumably monocistronic subgenomic mRNAs called sgRNAs (Fig. 16.9).
Fig. 16.9 : Genome of potexvirus; RsRp-RNA-dependent RNA polymerase; TGBp-triple gene block protein;CP-coat protein.
Replication of PVX:
Potexiras replicates inside the cytoplasm and it does not depend on a helper virus. Recent advances in potexvirus research have produced new models describing virus replication, cell-to-cell movement, encapsidation, R gene-mediated resistance and gene silencing. Virus penetrates the host cell, uncoates the virion and releases the genomic RNA into the cytoplasm.
The viral genome encodes structural proteins and non-structural proteins. Virions consist of 1 structural protein(s). Proteins constitute about 94% of the weight of a viral particle. The viral RNA is translated as a monocistronic mRNA to produce the RdRp (encoded by the 5′-proximal ORF).
Consequently, a (-) sense complementary ssRNA is synthesized using the genomic RNA as a template. Internal subgenomic promoters are used to transcribe the subgenomic RNAs (sgRNAs). Transduction of these sgRNAs yields the capsid protein (CP) and movement proteins (MP). The CP proteins and (+) sense RNA are assembled resulting in formation of new virus particles.
During replication, interaction between distant RNA elements is a central theme in potexvirus replication. The 5′ non-translated region (NTR) regulates the synthesis of genomic and subgenomic RNA and encapsidation, as well as transport of virus through plasmodesmata. The 3′ NTR regulates the synthesis of both (+) and (-) strands of RNA.
The intra- and inter-cellular transport of potexvirus requir interactions among viral RNA, CP and elements of the triple gene block proteins (TGBps). RNA sequence has a ‘triple gene block sequence’ (TGB) which encodes proteins involved in cell- to-cell movement. Possibly, species-specific interactions among movement proteins are obligatory for cell-to-cell movement of potexvimses.
How do the TGBps interact for virus movement is still unclear. As the TGBpl protein of PVX gates plasmodesmata, regulates virus translation and is a suppressor of RNA silencing more research is needed to determine how these properties contribute to propelling virus through the plasmodesmata. Specifically, TGBpl suppressor activity is required for virus movement, but how the silencing machinery relates to plasmodesmata is not known.
The TGBp2 and TGBp3 proteins are endoplasmic reticulum (ER)-associated proteins required for virus movement. TGBp2 associates with ER-derived vesicles that traffic along the actin network. So far it is unclear whether the virus-induced vesicles are cytopathic structures regulating events along the BR or are vehicles carrying virus to the plasmodesmata for transfer to neighbouring cells.
A single-tailed particle (STP) has been identified that comprises of RNA, coat protein (CP) and TGBpl. It has been proposed that TGBpl aids in transport of virions or single-tailed particle (STP) between cells and ensures translation of RNA in the receiving cells.
Epidemiology of PVX:
PVX spreads through rubbing, contact of plants and tubers, seed cutting knife, farm implements, clothing and animal fur. In stores it can spread by sprout contact also. Root clubbing may also transmit PVX to a limited extent. In nature a wide range of PVX-infected plant species including weeds (Dahlia, Solanum chacoense) serve as the source of virus infection.
Transmission of PVX:
A fungus Synchytrium endobioticum causing potato wart also transmits through oospores. Like TMV, it is also sap transmitted and no vector has been reported so far. It spreads through contact between the healthy and diseased plant, core grafting and dodder (Cuscuta compestris). However, it perennates in the diseased seed stocks. Average tuber infection varies from 13 to 23%.
Yield Loss in PVX:
PVX causes 10-30% yield loss. But the combined effect of PVX and PVY or PVA causes severe disease and reduce yield loss drastically. PVX-infected plants turn more or less susceptible to both early blight (Alternaria solani) and late blight (Phytophthora infestans) due to secretion of antifungal substances in leaves.
Extracts and washings from PVX-infected plant leaves suppressed the germination of A. solani conidia due to the presence of pathogenesis-related proteins (PRP). A comparative assessment of yield loss due to different potato viruses and viroids is given in Table 16.2.
Table 16.2 : Yield loss due to potato viruses/viroids.
3. Potato Virus Y (PVY):
PVY is also known as Solarium virus 2, potato virus 20 and potato leaf drop streak virus. It is distributed throughout the world on a wide range of hosts in Solanaceae, for example tomato, potato etc. Several strains of PVY have also been reported which differ in physical features. The thermal death point is 52°C for 10 minutes and longevity 24-48 hours at the laboratory conditions.
Symptoms of PVY:
Symptoms caused by PVY vary according to environment and host varieties. It ranges from weak mosaic to foliage necrosis. The common symptoms are leaf drops streak or acropetal necrosis. Secondary infected plants show less necrosis but are dwarfed with brittle, crinkled and branched leaves. In combination with PVX, it causes rugose mosaic of potato. This disease causes a severe damage to the plants.
It shows variable symptoms in different varieties of potato. The foliage is mottled which in turn becomes wrinkled, punctured and remarkably reduced in size too. The margin of leaflets gets rolled downwardly and plant is stunted.
Abnormal hairiness of leaves and dwarfing of whole plant occur. When the plants are severely infected they die before producing tubers. Host range of PVY is mainly Solanaceae but the virus also causes disease in Chenopodiaceae and Leguminosae.
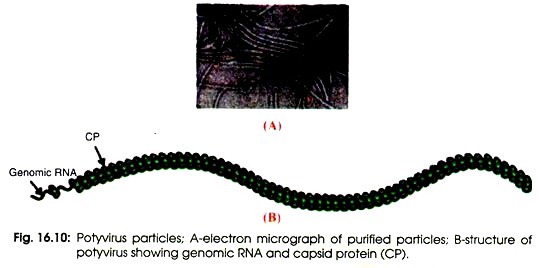
Structure of PVY:
The virions of a potyvirus consist of a non-enveloped capsid with helical symmetry. Virion is flexuous, filamentous, 720-850 nm long and 12-15 nm in diameter (Fig. 16.10 A). The axial canal can be distinct or indistinct but 2.5-3 nm in diameter. The basic helix is obvious or obscure and the pitch of the helix is 3.3-3.4 nm.
The definitive morphological structure is composed of approximately 2000 copies of capsid protein (CP) (B). Characteristic inclusion bodies are found within the infected plant cells. It is cytoplasmic virus but some have nuclear inclusions. It is not persistent in aphid vectors. It is the major virus of potatoes because it spreads easily and reduces the yield.
There are three types of strains of PVY viz., PVY° (common strains), PVNn (tobacco vein necrosis strains), PVYC (stipple streak strains). These strains are identified according to severity of symptoms in tobacco, potato, etc.
PVY is divided into three subgroups:
(i) Potyvirus (aphid-transmitted virus),
(ii) Bymovirus (fungus-transmitted virus), and
(iii) Rymovirus (mite-transmitted virus).
Genome Organization of PVY:
The R genome of potyvirus is mnonopartite and contains only one linear, (+) sense single-stranded RNA. The genome is sequenced and the complete sequence is about 9.000-10,080-12,000 nucleotides long. The genome has a base ratio of 21-23.51-26% guanine; 23-30.15-44% adenine; 14.9-22.41-28% cytosine; 15.6-24.41-30.9% uracil.
The 5′-end of the genome has a genome-linked protein (VPg). The 5′ non-coding region functions as an enhancer of translation. The 3′-terminus has a poly (A) tract and the genome has an intergenic poly (A) region. The genome organisation of a typical member is shown in Fig. 16.11 which indicates 10 mature proteins and the nine cleavage sites (see arrow).
The potyvirus contains one long open reading frame (ORF) which encodes one large polyprotein precursor of 350 kDa that is subsequently processed by 3 different virus-encoded proteases (all encoded by the virus itself) into 10 different mature functional proteins denoted as proteinase P1, helper component (HC), proteinase P3, cylindrical inclusion (CI), nuclear inclusion A (NIa), nuclear inclusion B (Nib), capsid protein (CP), as well as two small putative proteins known as 6K1 and 6K.2.
There are conserved and variable regions within the potyvirus genome. The conserved regions incorporate the helper component proteinase (HC-Pro) and nuclear inclusion B (Nib), whereas the variable regions consist of P1. P3, and the coat protein (CP). It has been reported that P3 displays low homology between species but despite the variation observed between P3 proteins of distinct poty viruses.
Two proteinases [P1 and the helper component proteinase (HC-Pro)] catalyse only auto-proteolytic reactions at their respective C termini. The remaining cleavage reactions are catalysed by the small nuclear inclusion protein (Nia-Pro). Many of the proteins have multiple functions. Dougherty and Carrington (1988) have described the expression of potyvirus proteins and their functions (Table 16.3).
Replication of PVY:
Virus penetrates into the host cell. Uncoating and release of the viral genomic RNA occur into the cytoplasm. The virion RNA is infectious and serves as both the genome and viral messenger RNA. The genomic RNA is translated into poly-proteins which are subsequently processed by the action of three viral-encoded proteinases into functional products.
The viral genome replicates within the cytoplasm; hence replication is cytoplasmic one. During transcription, the sub- genomic RNA is absent from infected cells. Virions may provide helper functions to dependent virus during replication. The virion acts a helper for another virus.
A negative-sense complementary ssRNA is synthesized by using the genomic RNA as a template. Thereafter, new genomic RNA is synthesized by using the negative-sense RNA as a template. Consequently, new virus particles are formed after assembly of genomic RNA and coat protein.
Yield Loss of PVY:
PYV is the most important virus causing about 80% loss according to variety, environment and virus strains. PVY can cause complete failure of the crop. In combination with PVS or PVA, PVY induces synergistic symptoms called rugose and crinckle mosaic, respectively. PYV infected plants are more susceptible to early and late blights.
Control of PVY:
A large number of varieties having high degree of resistance to PVY have been bred using Solarium tuberosum and S. stoloniferum. Six resistant genes have been identified in S. tuberosum.
4. Cucumber Mosaic Virus (CMV):
Cucumber mosaic virus (CMV) belongs to the genus Cucumovirus in the family Bromoviridae of the Group IV (+) ssRNA viruses. This family consists of several genera Alfamovirus Anulavirus, Bromo-virus, Cucumovirus, Ilarvirus and Olea-virus.
Cucumber mosaic virus is a species of the genus Cucumovirus. CMV occurs in several countries including India. It infects about 100 plants belonging to about 20 families. It occurs most frequently on the members of Cucurbitaceae, tomato, potato, carrot, spinach, etc.
Symptoms of CMV:
Leaf mottling, crinkling and curling of edges occur on cucumber. Petioles and internodes are shortened resulting in stunted and compact appearance of the plant. If the plants are infected early in season, flowers do not develop, and if infected late only a few small sized grains are formed. The fruits too are mottled with light yellow and white patches. The older leaves eventually dry. On the other plants symptoms differ.
Structure of CMV:
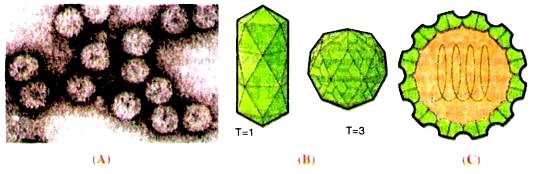
Virions consist of a capsid which is non-enveloped, icosahedral or bacilliform 26-35 nm in diameter. Symmetry is icosahedral (T = 3 or T = 1) (Fig. 16.12). The three genomic and one sub-genomic segments are encapsidated in distinct particles, resulting in several different types of virion.
Virion consists of one structural protein present in the capsid. The arrangement of capsomers is not obvious, possibly capsid consists of 32 capsomers. Virus preparations contain more than one particle component.
Fig. 16.12 : Virons of cucumber mosaic virus; A-an electron micrograph; B-a bacilliform virion (T=1) and an icosahedral (T=3) virion; C-structural organization of a virion having ssRNA geneome.
The viral genome encodes both structural proteins and non-structural proteins. The thermal inactivation point (TIP) is at 55-70°C and longevity in vitro (LIV) is 1-10 days. Its infectivity does not change after treating it with ether, or deproteinizing with proteases, phenol or detergent.
The virion consists of one (+) ssRNA genome which genome is segmented, tripartite (segments are distributed among three particle types of different size), and consists of 4-5 segments of linear positive-sense, single-stranded RNA (Fig. 16.12 C).
Genome of CMV:
Genome is segmented, tripartite linear (+) sense ssRNA which is composed of RNA1, RNA2 and RNA3. A 3′ tRNA-like structure and a 5’cap are present on each genomic segment of RNA (Fig. 16.13). Genomic nucleic acid is infectious i.e. it does not require coat protein or sub-genomic mRNA-4 for infection. Minor species of non-genomic nucleic acid are also found in virions.
The encapsidated nucleic acid is mainly of genomic origin, but virions may also contain sub-genomic RNA and satellite RNA. In some isolates virions contain mRNA derived from genomic RNA-3. The complete genome is 8,621 nucleotides long. RNA-1 is fully sequenced and complete sequence is 3,389 nucleotides long. RNA-4 is sub-genomic which has been sequenced and the complete sequence is 1,000 nucleotides long.
The 5′-end of the genome (on all 4 RNAs) has a cap of methylated nucleotides having a sequence m7G5’ppp5. The 3′ end has no poly (A) tail; instead the 3′ terminus has conserved sequences of 200 nucleotides, which has an amino-acylated tRNA-like structure. Moreover, the genome has no poly (A) in the intergenic region.
The multipartite genome is divided into 3 to 4 different types of particles; the largest particles contain each one molecule of RNA-1, the medium sized particles contain each one molecule of RNA-2, and the smallest particles contain one molecule each of RNA-3 and RNA-4.
Replication of CMV:
Viral replication is cytoplasmic; moreover, it does not depend on a helper virus. The molecular receptors on host cell surface play a role in recognition of virions. Virus attaches to host receptors, un-coats the capsid, and releases the genomic RNA into the cytoplasm. RNA1 and RNA2 encode protein la and protein 2a, respectively.
Both the proteins promote the replication of genomic RNAs and internal transcription of sgRNA4 from the minus-strand copy of RNA3. Consequently, a (-) sense complementary ssRNA is synthesized by using the genomic RNA as a template. New genomic RNAs are synthesized by using the (-) sense RNAs as templates.
Sub-genomic RNA4 is transcribed from RNA3 anti-genome. Sub-genomic RNA4 is translated to produce capsid proteins. New virions are assembled after completion of protein synthesis. Finally virions are released by lysis of the infected host cell.
5. Cauliflower Mosaic Virus (CaMV):
Cauliflower mosaic virus (CaMV) is a type member of the caulimoviruses which falls under the family Caulimoviridae in the Group VII dsDNA-RT viruses. The CaMV are the only plant viruses that have dsDNA genome.
Due to the presence of dsDNA, the caulimoviruses are exploited as vector in genetic engineering of plants. The CaMV consists of open circular dsDNA as genetic material with single strand discontinuity like hepadna Virus. The DNA is linear in situ but gets circularized after extraction. CaMV shows icosahedral symmetry of the capsid with a 50 nm diameter. It consists of more than one protein shell.
In theiytoplasm of infected cauliflower leaves, CaMV forms characteristic X bodies which are rounded structure:
Symptoms of CaMV:
From the cell walls of infected leaves, there arise finger like processes. It has also been observed that mitochondria and nuclei become abnormal in the infected cells of host leaves.
Structure of CaMV:
Virions occur in the cytoplasm, and in some cases in the nucleus. Cytoplasmic virions are associated with electron-dense proteinaceous inclusion bodies (for caulimoviruses, the product of ORF6). The product of 0RF2 also forms inclusion bodies which are electron translucent.
Inclusion bodies can be seen by light microscopy as well as by electron microscopy. The virion CaMV shows an icosahedral symmetry with a diameter of 52 nm which is made up of 420 capsid protein (CP) subunits arranged with a triangulation T = 7 (Fig. 16.14).
The protein subunits surround a solvent-filled central cavity. Each virion consists of a circular dsDNA molecule of about 8.0 kb which is interrupted by site-specific discontinuities.
It results from its replication by reverse transcription. The single stranded nicks in the viral DNA are repaired after entering the host. It results in formation of a supercoiled molecule that binds to histone proteins. Transcription of this DNA results into a full length 35S RNA and a sub-genomic 19S RNA molecule, which are terminally redundant.
Genome of CaMV:
The genome is a mono-partite, open circular dsDNA of about 8000 base pairs with one discontinuity in one strand, and one or more discontinuity in the other strand (Fig. 16.15).
Strands displacement causes gaps in the genome, which are associated with the replication of the virus. The promoter of the 35S RNA is very strong. It is used in plant transformation. The 35S RNA is complex and contains one leader sequence (600 nt long). This leader is followed 6 to 8 tightly arranged longer ORFs that encode all the viral proteins.
There are six major coding regions (I, III, IV, V, VI, VI11) and 2 minor coding regions (II. VII) in the genome. The minor coding regions act as the store house of the non- essential genes. The coding regions of ORFs and their function are given in Table 16.4. The mechanism of expression of these proteins is very special.
The ORF VI protein (encoded by the 19S RNA) controls translation re-initiation of major open reading frames oh the polycistronic 35S RNA, which results in release of virions. After making association with polysomes and eukaryotic initiation factor eIF3, TAV starts its function.
Replication of CaMV:
CaMV replicates by reverse transcription in cytoplasm/nucleus. Virion of CaMV interacts with a cellular receptor of the host and enters inside the cell. The viral dsDNA genome is transported to the nucleus where the host RNA polymerase II carries out its transcription.
Then translation of 35S RNA and 19S RNA takes place where viral proteins are produced thereafter, reverse transcription of 35S RNA occurs to produce new dsDNA genomes in cytoplasm. Capsid protein encapsulates the viral genome. The new viral particles get targeted to inclusion body and are released outside.
6. Potato Leaf Roll Virus (PLRV):
Potato leaf roll virus (PLRV) belongs to the Genus Polerovirus, family Luteoviridae and Group IV (+) sense ssRNA virus according to the Baltimore classification.
The family Luteoviridae consists of three genera viz., Polerovirus, Luteovirus and Enamovirus. Leaf curl of tomato has been reported from many parts of the world. It is most common is winter season in India. It has non-enveloped particles with isometric symmetry. It possesses one linear (+) ssRNA as genetic material.
The viral host range is restricted to members of family Solanacae. PLRV is transmitted via green peach aphids (Myzus persicae) in a circulative manner without any evidence for propagation in its invertebrate vector.
It is restricted to the phloem tissue of the infected plants:
Symptoms of PLRV:
The symptoms that develops in the infected plants are dwarfing, puckering, twisting and curling towards the dorsal side of leaves, mottling, vein clearing, excessive branching, shortening of whole plant, and partial or complete sterility in the plants.
Structure of PLRV:
PLRV particles are isometric icosahedral, non-enveloped virion (T=3 particles) of 25-30 nm diameter with 180 copies of the capsid protein (CP) (Fig. 16.16). It contains 30% of single stranded, positive-sense RNA with a molecular weight of -5.9 kb. The viral genome is covalently linked to a 7.2 kDa protein (VPg) at its 5′ start and not polyadenylated at the 3’end.
Genome Organization of PLRV:
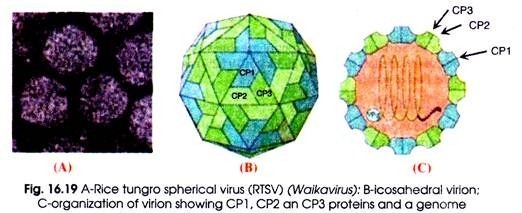
Virion consists of a linear, (+) sense ssRNA genome of 5.3-5.7 kb in size (Fig. 16.19). Poleroviruses have a VPg bound at the 5′ end. There is no poly (A) tail or tRNA-like structure at the 3′ end. Different isolates of PLRV have been sequenced. The RNA codes for at least 8 open reading frames (ORFs) that are located in two clusters of genes and separated by 197 nucleotides (nt) of non-coding sequences.
The first cluster is preceded by 1 74 nt of non-coding sequences, whereas the second cluster is followed by 141 nt of non-coding sequences. The first gene cluster is translated directly from genomic RNA to produce ORFO (with Mr of 28 kDa and responsible for the symptom development), ORF1 (with Mr of 70 kDa with a chymotrypsin-like serine protease domain), and the VPg (downstream of the protease domain).
ORFO and ORF1 are overlapped in different frames. ORF2 (with Mr of 118 kDa) is produced as a frame-shift from ORF1 and contains RNA dependent RNA polymerase (RdRp) motifs.
The only proteins translated directly from genomic RNA are the polymerase (ORF2) which is expressed as a fusion protein by – 1 translational frameshift at the end of ORF1, and ORF1 protein. All other ORFs are translated from sub-genomic RNAs (sgRNAs). The second cluster is translated from sub-genomic RNA1 (sgRNA1) and sgRNA2 with Mr of 2.3 kb and ~0.8 kb, respectively.
The sgRNA1 serves as an mRNA [for the coat protein (ORF3)] with MW of 23 kDa and completely overlaps the 17 kDa movement protein (MOVP) (ORF4) but in a different frame and the aphid transmission factor (ORF5) that is located directly downstream the ORF3 amber stop codon and encodes a 53-kDa polypeptide.
The sgRNA2 serves as mRNA for at least two ORFs. ORF6 present within the C terminus of ORFS but in other frame with Mr of 7.1 kDa and ORF7 representing the C terminus of ORFS with Mr of 14.1 kDa with a proposed transcriptional regulation function. Read-through of the CP stop codon produces CP-ORF5 (CP extended) i.e. the capsid subunit essential for aphid transmission (Fig. 16.17).
Fig. 16.17 : Genome organisation and gene expression of Polerovirus
Replication of PLRV:
Polerovirus replicates inside the host’s cytoplasm and it does not depend on a helper virus. The virion RNA is infectious and serves as both the genome and viral messenger RNA.
Virus particle penetrates into the host cell, un-coats, and releases the genomic RNA into the cytoplasm. The viral RNA ORF1 and ORF2 are translated to produce the RdRp fusion protein. A negative-sense complementary ssRNA is synthesized by using the genomic RNA as a template.
Thereafter, new genomic RNA is synthesized by using the negative-sense RNA as a template. The negative-sense complementary ssRNA also serves as template for the synthesis of 3′ co-terminal sgRNAs. Transduction of these sgRNAs yields the capsid (and extended CP) and movement proteins. Consequently formation of new virus particles takes place.
Transmission of PLRV:
The green peach aphid, Myzus persicae is also one of the most effective vectors. During winter, the virus remains within the infected plant and acts as the primary source of the PLRV inoculum. After infection the virus is largely contained to the phloem tissue and infects daughter tubers through the vascular system.
Control of PLRV:
So far no definite control measures have given fruitful result. However, developing resistant varieties, spraying with Ekatox (0.02%) and Regor (0.05%) at 10 days interval reduce the infection of plants.
7. Mosaic Disease of Sugarcane:
Sugarcane is one of the important crops of India which has been sown since the dawn of human civilisation. Besides the attack of several pathogens and pests, it is infected by many viruses which cause different types of diseases such as Fiji disease, chlorotic streak, mosaic, ratoon stunting, Australian dwarf, etc.
In 1921, this disease was noticed first. But it is widespread in all sugarcane growing areas of India. It becomes a potential threat through development of pathogenic strains.
Mosaic of sugarcane is caused by sugarcane mosaic virus (SCMV) or Saccharum virus 1 or sugarcane virus 1 that falls under the Group IV (+) sense ssRNA viruses. It is a member of family Potyviridae. There are six different strains of SCMV that cause mosaic disease.
For the development of symptoms, SCMV must be inside the living tissues especially in phloem established bundles. Six weeks after planting there develop pale patches or blotches in the green tissues of the leaves.
Moreover, the diseased leaves show mottling, chlorotic or light coloured, irregularly spread stripes or streaks. An area of proteolysis develops in one part of the cytoplasm in the cells of affected plants. This areas shows a vacuolated mass having X bodies. These can be heavily stained and observed under microscope.
There are three principal modes of spread of SCMV: by aphid vectors, infected seed cane, and mechanical inoculation. Only aphid vectors and infected seed cane are important in the field.
Mechanical transmission i important only in greenhouse and laboratory. There are at least 12 species of aphids that can transmit SCMV from diseased to healthy sugarcane. Perennial grasses also act as host and a source of reservoir of inoculum of SCMV.
There are several methods which have been recommended for the control of mosaic disease of sugarcane such as:
(a) Use of healthy setts as seeding material,
(b) Systemic roguing of infected cane when infection is not severe,
(c) Elimination of perennial grasses acting as weeds and host,
(d) Avoiding rationing practices if plants are heavily infected,
(e) Using resistant varieties,
(f) Operating of good quarantine, etc..