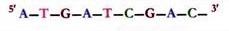An exclusive project report on DNA. This project report will help you to learn about: 1. Introduction to DNA 2. Constituents of DNA 3. Molecular Structure 4. Forms 5. DNA as the Genetic Material 6. DNA Content 7. Unique and Repetitive DNA.
Contents:
- Project Report on Introduction to DNA
- Project Report on the Constituents of DNA
- Project Report on the Molecular Structure of DNA
- Project Report on the Forms of DNA
- Project Report on DNA as the Genetic Material
- Project Report on DNA Content
- Project Report on Unique and Repetitive DNA
Project Report # 1. Introduction to DNA:
The chromosomes are made up of two types of macromolecules — proteins and nucleic acids. The nucleic acids are of two types, viz. deoxyribonucleic acid (DNA) and ribonucleic acid (RNA). DNA and RNA are chain-like macro-molecules that function in the storage and transfer of genetic information.
They are major components of all cells. DNA is found predominantly in the nucleus while RNA is predominant in cytoplasm. DNA is the genetic material of most organisms including many viruses. Some viruses,’ however, have RNA as their genetic material.
Deoxyribonucleic acid (DNA) is found in the cells of living organisms except plant viruses. DNA is double stranded but in some cases like bacteriophages (e.g., φ x 174) the DNA is single stranded and remains coiled and enclosed in a protein coat. DNA may be circular in bacteria, spirally coiled and un-branched threads in mitochondria and plastids of eukaryotic cells.
Project Report # 2. Constituents of DNA:
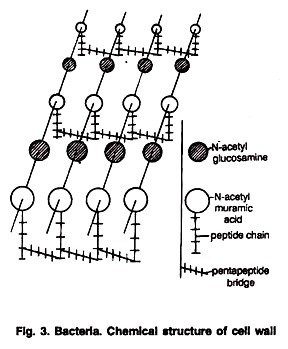
Deoxyribonucleic acid is a long chain polymer (polynucleotide) composed of monomeric units, called nucleotides. Each nucleotide is composed of a nucleoside (a sugar and a base) and a phosphate group.
Chemical analysis of highly purified DNA have shown that it is made of four kinds of monomeric building blocks each of which contains three types of molecules :
(i) Phosphoric acid
(ii) Sugar molecule, and
(iii) Organic bases.
(i) Phosphoric Acid:
The phosphoric acid (H3PO4) in the nucleic acid is called phosphate (Fig 4.1). Phosphoric acid has three reactive hydroxyl (-OH) groups of which two are involved in forming sugar phosphate backbone of DNA. The phosphate makes a nucleotide negatively charged.
(ii) Sugar Molecule:
DNA contains a five carbon sugar, hence it is a pentose sugar (Fig. 4.1). Since one oxygen atom at the 2′ carbon is missing, hence it gets its name 2′-deoxyribose. Four of the five carbon atoms plus a single oxygen atom forms a five-membered ring. The fifth carbon atom is outside the ring and forms a part of a -CH2 group.
(iii) Organic Bases:
Different types of heterocyclic nitrogen containing ring compounds are found in the structure of DNA. They are called simply as bases because they can combine with H^ in acidic solution. They are also referred to as nitrogenous bases due to presence of nitrogen.
The bases are of two types mainly — Pyrimidine and Purine.
(a) Pyrimidine:
Pyrimidine bases are made up of a six-membered pyrimidine ring which is similar to the benzene ring except that it contains nitrogen in place of carbon at the positions 1 and 3. Pyrimidine bases are of 2 types — thymine and cytosine (Fig. 4.1), commonly abbreviated as T and C respectively.
(b) Purine:
Purine is a derivative of pyrimidine. It consists of a pyrimidine ring and a five- membered imidazole ring (having nitrogen at 7 and 9 positions) which are fused together at 5 and 4 positions. There are two purine compounds namely – adenine (A) and guanine (G) (Fig. 4.1).
Rare or minor bases:
In addition to the four common bases (A, T, G, C), certain other unusual bases of purine and pyrimidine derivatives, called rare or minor bases, occur in small amounts in DNA of some organisms.
In some viruses, uracil occurs in place of thymine in DNA. The T-even phages contain 5-hydroxy- methyl-cytosine in place of cytosine. These modifications protect the viral DNA from degradation by the host cell endonuclease. Other rare bases in DNA are 5-methyl-cytosine, N6-methyl- adenine, N2-methyl-guanine, etc.
Molar Ratio of Nitrogenous Bases in DNA (Chargaff Rules, 1955):
(i) The purine and pyrimidine components occur in equal amounts in a DNA molecule.
(ii) The amounts of adenine (A) is equivalent to the amount of thymine (T) and the amount of cytosine is equivalent to that of guanine (G).
(iii) In DNA, A + G / T + C value is always one or nearly one.
(iv) The base ratio A +T/G + C may vary in the DNA of different groups of organisms but is constant for particular species. Therefore, this ratio has been used to identify the DNA from a particular species.
Project Report # 3. Molecular Structure of DNA (Watson and Crick’s Model):
In 1953 Watson and Crick postulated a three dimensional working model of DNA, i.e., double helix structure of DNA based on the X-ray data of Wilkins and base-equivalence observed by Chargaff.
Nucleoside:
A base combined with a sugar molecule is called a nucleoside. When deoxyribose sugar binds with base, it makes deoxyribonucleoside. Obviously, in DNA four different nucleosides are found.
These are:
(i) Deoxycytidine;
(ii) Deoxythymidine;
(iii) Deoxyadenosine; and
(iv) Deoxyguanosine.
Nucleotide:
A nucleotide is derived from a nucleoside by addition of a molecule of phosphoric acid. The phosphate molecule is linked with sugar molecule at carbon number 5 or at carbon number 3 (Fig. 4.2).
The nucleotides in DNA are of 4 types:
(i) Deoxycytidylic acid;
(ii) Deoxythymidylic acid;
(iii) Deoxyadenylic acid;
(iv) Deoxyguanylic acid (Fig. 4.3, Table 4.1).
Polynucleotide:
A number of deoxyribonucleotides are covalently linked one by one to form a polynucleotide chain, i.e., deoxyribonucleotide monomer units are united through the formation of phosphodiester bonds (a diester bond is one which involves two ester bonds). Fig. 4.4.
Double Helix:
Watson and Crick suggested that in a DNA molecule, there are two such polynucleotide chains which are coiled about one another in a spiral.
The two polynucleotide strands are held together in their helical configuration by hydrogen bonding between bases in opposing strands, the resulting base pairs being stacked between the two chains perpendicular to the axis of the molecule like the steps of a spiral stair case (Fig. 4.5).
The base-pairing is specific; adenine is always paired with thymine, and guanine is always paired with cytosine (Fig. 4.6). Thus, all base-pairs consist of one purine and one pyrimidine.
The specificity of base-pairing results from the hydrogen-bonding capacities of the bases in their normal configurations. In their most common structural configurations, adenine and thymine form two hydrogen bonds, guanine and cytosine form three hydrogen bonds (Fig. 4.7).
The two strands of DNA double helix are thus said to be complementary (not identical). This property, that is complementarity of the two strands, makes DNA uniquely suited to store and transmit genetic information. The base-pairs in DNA are stacked 3.4 Å apart with 10 base-pairs per turn (360°) of the double-helix.
The sugar- phosphate backbones of the two complementary strands are antiparallel, that is, they have opposite chemical polarity.
As one moves unidirectionally along a DNA double helix, the phosphodiester bonds in one strand go from a 3′ carbon of one nucleotide to a 5′ carbon of the adjacent nucleotide, whereas those in complementary strand go from a 5′ carbon to a 3′ carbon. This opposite polarity of the complementary strand is very important in considering the mechanism of replication of DNA.
The high degree of stability of DNA double helices results in part from the large number of hydrogen bonds between the base-pairs and in part from the hydrophobic bonding between the stacked base-pairs. The planar sides of them are relatively nonpolar and thus tend to be water insoluble (hydrophobic).
This hydrophobic core of stacked base-pairs con- tributes considerable stability to DNA molecules present in the aqueous protoplasm’s of living cells.
Project Report # 4. Forms of DNA:
The DNA molecules exhibit a considerable amount of conformational flexibility. It can exist in A, B, C, D and Z forms (Table 4.2). B form (B-DNA) is the structure proposed by Watson & Crick and is the native conformation of DNA in solution.
It consists of a right-handed antiparallel double helix of sugar-phosphate backbone, with purine-pyrimidine base-pairs roughly perpendicular to the axis of the helix. The tilt of base-pairs of the helix is 6.3°. One turn of the helix consists of 10 base-pairs. The rise of the helix per base pair is 3.37 Å.
A form (A-DNA) has 11 base pairs. The base pairs are considerably tilted from the axis of the helix. The axial rise is 2.56Å. The helix observed under conditions of dehydration and high concentrations of salt is wider and shorter than B- helix, the distinction between the major and minor grooves are reduced.
C form (C-DNA) results by reduction of hydration of the B form below 66% with excess of salt still present. The size of helix of C form DNA is greater than Å type of DNA but is smaller than B-DNA. It is about 31 Å. There are 9.33 base pairs per turn. The axial rise of base pairs is 3.32 Å with a tilting of about 7.8°.
D form (D-DNA) and E form (E-DNA) are found rarely as extreme variants. In case of D- form there are 8 base pairs per turn of helix. An axial rise of base pairs is 3.03 Å with tilting of about 16.7°.
In case of E-form, there are 7.5 base pairs per turn of helix. Z form (Z-DNA) is an unique left-handed (Fig. 4.8) double helical form with a zig-zag (Fig. 4.9) sugar-phosphate backbone in antiparallel organization. This DNA has been called Z-DNA.
Project Report # 5. DNA as the Genetic Material:
Transformation Experiment:
Transformation experiment was initially conducted by F. Griffith in 1928 (Fig. 4.21). The injected a mixture of two strains of Pneumococcus (Diplococcus pneumoniae) into mice. One of these two strains, S III was virulent and other strain R II was non-virulent (causing no infection).
Heat-killed virulent S III strain when injected, showed that infectivity after heat killing is lost. The mice injected with a mixture of R II (living) and S III (heat killed) died and virulent Pneumococcus could be isolated from these mice. This phenomenon was described as transformation.
O. T. Avery, C. M. Macleod and M. McCarthy repeated Griffith’s experiment in an in vitro system in order to identify the transforming principle responsible for converting non-virulent into virulent type and reported their results in 1944 (Fig. 4.22). Virulence in Pneumococcus depends on a polysaccharide capsule which is present in virulent strain S III and is absent in non-virulent strain R II.
The cells of non-capsulated type – Rll were treated with an extract of DNA from capsulated strain S III. A few cells of S III type could be isolated from the mixture.
This phenomenon of transferring characters of one strain to another by using a DNA extract of the former is called transformation. When the extract was treated with DNAse (an enzyme which destroys DNA) this transforming ability was lost. Proteases (enzymes which destroy proteins) did not affect the transforming ability. These experiments thus indicated that DNA and not the protein, is the genetic material.
Hershey-Chase Experiment:
Additional direct evidence indicating that DNA is the genetic material was published in 1952 by A. D. Hershey and M. Chase. The experiment showed that genetic information of a particular bacterial virus (bacteriophage T2) was present in DNA. Bacteriophage T2 infects the common colon bacillus E. coli (Fig. 4.23).
The basis for Hershey-Chase experiment is that DNA contains phosphorus but no sulphur, whereas proteins contain sulphur but no phosphorus.
Thus, Hershey and Chase were able to specifically label either (1) the phage DNA by growth in a medium containing radioactive isotope of phosphorus 32P, in place of the normal isotope 31P, or (2) phage protein coats by growth in a medium containing radioactive sulphur 35S, in place of the normal isotope 35S (Fig. 4.24).
When T2 phage particles labelled with 35S were mixed with E. coli cells for a few minutes and were then subjected to shearing forces by placing the infected cells in a warring blender, it was found that most of the radioactivity (and thus the proteins) could be removed from the cells without affecting progeny phage production.
When T2 phage in which DNA was labelled with 32P were used, essentially all radioactivity was found inside the cells, that is, it was not subjected to removal by shearing in a blender (Fig. 4.25).
The sheared off phage-coats were separated from the infected cells by low-speed centrifugation which pellets (sediments) cells while leaving phage particles suspended. The results indicated that DNA of the virus enters the host cell, whereas protein coat remains outside the cell.
Since progeny viruses are produced inside the cell, Flershy and Chase’s results indicated that the genetic information directing the synthesis of both the DNA molecules and protein coats of the progeny viruses must be present in the parental DNA. Moreover, progeny particles were shown to contain some of the 32P, but none 35S.
Project Report # 6. DNA Content:
C-Value Paradox:
Large variation in DNA content among species of the same genus as well as among genera of the same family has been observed. This wide range of variation in the nuclear DNA contents among species within a genus may be partly attributed to differences in chromosome numbers.
However, species at the same ploidy level and with the same chromosome number may either show little or ‘no variation (e.g., Hordeum, Avena, etc.) or may exhibit many-fold differences (e.g., Lathyrus, Vicia, Helianthus, Crepis, Allium, etc.). This inter specific variation may be continuous or discontinuous.
Intra- specific variation in DNA content has also been reported in recent years, so that the variation of DNA among genotypes is a rule rather than an exception. This variation has sometimes been used to explain mechanism of evolution of specific groups.
In general, the huge difference in DNA amount not necessarily correlated with the nature and status of the organism, is principally due to the repetitive DNA sequences present in higher amount. In the plant system, nearly 70- 80% of the DNA in the cereals is repetitive and only a fraction of the DNA controls the structural genes for qualitative characters.
In fact, the comparison of the genomes of different plant species ranging from algae to angiosperms show marked variation in the C value.
The haploid un-replicated DNA content of an individual is described as its C value. C value is nearly of 600-fold difference within the angiosperms alone, ranging from 0.2 pg in the crucifer – Arabidopsis thaliana to 127pg in Fritillaria assavrica, a liliaceous species. This paradoxical situation in C value termed as C value paradox is principally attributed to repetitive DNA content.
In human, of the 3 billion base pairs which constitute the entire genome, only about 50000 genes are supposed to be present. More than 40% of the DNA is highly repetitive. In general, only a fraction of the total amount of DNA codes for structural proteins and enzymes, the rest are repeats – noncoding or coding for non-specific-effect.
Project Report # 7. Unique and Repetitive DNA:
The chromosomes of prokaryotes contain DNA molecule with unique (non-repeated) base-pair sequences, i.e., each gene which is a linear sequence of few thousand base-pairs, present only once in the genome. If prokaryotic chromosomes are broken into many short fragments, each fragment will contain a different sequence of base-pairs.
In higher organisms on the other hand, the unique DNA sequences which are principally responsible for qualitative characters are present in much lower amount than that of the repetitive sequences. Such unique sequences are present in the chromosomes of higher organism in between repetitive sequences, controlling enzyme-protein.
The chromosomes of eukaryotes in general are very complex. Certain base sequences are repeated many times in the haploid chromosome complement, sometimes as many as million times. DNA containing such repeated sequences, called repetitive DNA, often representing a major component of the eukaryotic genome.
Repetitive DNA:
Types of Repetitive DNA:
The discovery of multiple copies of similar DNA sequences in chromosomes noted by Crick is a major event in the study of chromosome research. These repeats may be highly homogeneous, as in satellite DNA sequences in Xenopus, or may be moderate or minor in nature.
These sequences may also be inverted as in palindromes. In the chromosome structure, the highly homogeneous repeats may be tandem located in one locus in cluster form, whereas minor or moderate repeats may be interspersed located in intercalary positions or terminal. In tandem repeats, each sequence arranged adjacent to the other forming monomeric unit.
Tandem repeats are of two types — similar repeats and complex combinations of different repeats, interspersed repeats are highly scattered, i.e., dispersed throughout the genome along with other sequences. Dispersed repeats include mobile elements such as long interspersed nucleotide element (LINE), short interspersed nucleotide element (SINE), long terminal repeats (LTR).
Repeats may be microsatellites (simple sequence repeats), minisatellites, satellites.
Size of Repetitive DNA:
The length of repetitive sequences may vary from simple sequence repeats of di-, tri-, tetra- or hexanucleotides to nucleosome repeats of 180 bp and up to 10000 bp or more in rDNA repeats.
Amount of Repetitive DNA:
The demonstration of the repeated sequences accounts, to a great extent, for the huge amount of DNA, noted in the different organisms. In higher organisms only little amount of the DNA represents structural genes containing unique, the rest being amplification of non-coding sequences or repeats.
In the biological system, as a whole, the nuclear DNA content varies amongst the organisms without any concomitant increase in the number and structure of genes.
The importance of repetitive DNA sequences can be judged from the very fact that in the human system almost 40% of DNA is repetitive in nature. In several plant species, as a rule 72-75% of DNA is repetitive whereas in Drosophila only 50% constitutes the sequences.
Location of Repetitive DNA:
Repetitive DNA sequences are present throughout the chromosomes including in centromere and telomeres (also may be pericentromeric and sub-telomeric), introns, nucleolar organizing regions, segments of heterochromatin.
In general, in the entire chromosome structure, normally highly repeated or homogeneous repeats are located in one locus, e.g., secondary constriction or centromere and moderate, minor repeats are interspersed throughout. Highly homogeneous repeats have been located in ribosomal RNA (5S, 18S, 5.8S, 25S) gene loci and are mostly AT-rich.
A characteristic repeat (GGGGATT) found at the telomeres. Accessory (B) chromosomes which are heterochromatic in nature, rich in highly repeated sequences. Large portion of cereal genome is with interspersion of short repeats. Presence of repetitive sequence in introns, in transposons and in flanking regions of replicon has been noted.
Origin of Repetitive DNA:
The mechanisms suggested for origin of repeated sequences include salutatory replication, unequal crossing over, transposition including insertion. Repeated sequences are susceptible to change resulting from amplification, translocation, deletion and mutation leading to novel genomic configuration. Sequences in new environments may undergo patterning and amplification, leading to new repeat families.
Functions of Repetitive DNA:
Several non-specific functions involving cell-nuclear size and volume, chromosome cycle, generation time, duration of meiosis, chromatin folding have often been attributed to repeated sequences.
Role of interspersed repeats has been suggested for repair synthesis, regulation of chromosome structure in folding during pairing, acting as initiation points in replication, gene expression through methylation, gene conversion and compression.
Function attributed to palindromic repeats at different levels of protein synthesis includes recognition systems, both at DNA and RNA levels, involving deletion and translocation, cleavage sites, termination of transcription, binding of regulatory proteins and attachment of chromosomes with each other for information transfer.
Intron repeats facilitates alternate splicing or reshuffling of exons and inter-genic conversion, permit dispersion and promote genetic diversity through mobility. Accessory chromosome repeats in plants are associated with adaptation in different environmental set up.
Significance of Repetitive DNA:
Some of the simple sequence repeats can serve as good markers. Certain tandem repeated sequences may be unique to a particular species. Their distribution and copy number help in differentiating various linkage groups in the chromosome complement.
DNA fingerprinting and molecular hybridization involving repeats have immense potential in documentation and analysis of biodiversity, and tracing the trends in evolution. The sequence, location and frequency of short tandem repeats (STR), which is common in plants’ genome, have role in determining phylogenetic status of species.
Detection of Repeats Tm and Cot Value:
For detection of repetitive DNA, the double stranded DNA is first denatured by heating into single stranded DNA which is accompanied with increase in optical density (hyperchromicity).
The single stranded DNA is then allowed to cool slowly to cause re-association between the complementary sequences into double stranded DNA which accompanies decrease in optical density (hypochromicity). Tm is the temperature at which 50% re-association is achieved.
The formation of double stranded DNA is actually measured over different values of a parameter which is described as Cot value (conc. x time). If solution of different DNA concentrations is to be compared, same Cot value can be achieved by altering the time allowed for re-association.
For DNA sample with high proportion of repetitive DNA, re-association will be faster and higher degree of re-association achieved at lower Cot values.
Localization of Repeats:
For localization of repetitive DNA, chromosome banding and molecular in situ hybridization (ISH) techniques are being applied.
Chromosome Banding:
Differential banding patterns of chromosomes, usually observed at specific regions on particular levels, were initially developed for the analysis of human chromosome segments.
These bands are made visible through low and high intensity regions under the fluorescence microscope or as differentially stained areas under the light microscope. The methods were then extended first to different animals and later to plant chromosomes.
The protocol for molecular hybridization, that is, denaturation at the cytological level, if followed by renaturation and staining with different dyes, particularly Giemsa, gives intensely positive reaction at similar segments of chromosomes which otherwise show repetitive DNA.
Obviously such treatment is capable of revealing repetitive segments in chromosomes. This banding, following denaturation-renaturation and Giemsa staining, is termed G-banding.
Earlier, Caspersson and his colleagues had recorded differential fluorescence of different chromosome segments following staining with various fluorochromes (quinacrine) and observation under the ultraviolet microscope and had successfully employed it in formulating a banding pattern analysis of the human karyotype.
Such bands were referred to as Q-bands. Other fluorochromes produce banding patterns as well, e.g., Hoechst 33258 similar to Q and ethidium bromide the reverse.
These banding patterns are unique for each chromosome like fingerprints. The bands are generally consistent for a taxon except for minor variations. A number of other chemicals can also produce bands either identical with or different from the fluorescent ones, based on different principles.
Such differential banding patterns, usually observed at specific regions on particular chromosomes, are being increasingly used for the identification of chromosomes.
The chromosome band nomenclature, adopted at the Paris Conference in 1971, recognized the following types of banding in human chromosomes (Fig. 4.26A):
Q-bands:
By quinacrine staining and fluorescence.
G-bands:
By staining techniques using Giemsa and related stains after appropriate pre- treatment.
R-bands or banding reverse to Q-bands:
By staining with Giemsa after heating to 87°C.
C-bands:
For constitutive heterochromatin, demonstrated by the denaturation-re-association technique.
E-bands:
Produced by enzymic digestion, as classified by Lejeune (1973), show woolen and shrunken regions corresponding to the dark and faint regions of G-banding. The same nomenclature can be applied to banding patterns observed in other eukaryotic chromosomes.
Other banding patterns of chromosomes include:
CT-bands:
The centromeric and telomeric segments show bands after treatment in barium hydroxide, incubation and staining in ‘Stains All’ (4, 5, 4′, 5′-dibenzo-3, 3′-diethyl-9-methyl-thi- acarbocyanine bromide).
N-bands:
At nucleolus-organizing regions, possibly due to acidic proteins.
O-bands:
With orcein staining, mainly for plant chromosomes; both intercalary and centromeric heterochromatin show bands. All the above methods of banding bring out clearly the differentiation of chromosome segments which can easily be analysed under the microscope. This technique permits identification of chromosome segments with specific molecular complexity such as repeat sequence.
The comparison of banding pattern between different genotypes, altered and unaltered, can localize the segments which have undergone alterations. The importance of banding can be judged by the very fact that, R-banding that is the reverse banding has been utilized for mapping of gene sequences in human chromosome.
In Situ Hybridization (ISH: FISH & GISH):
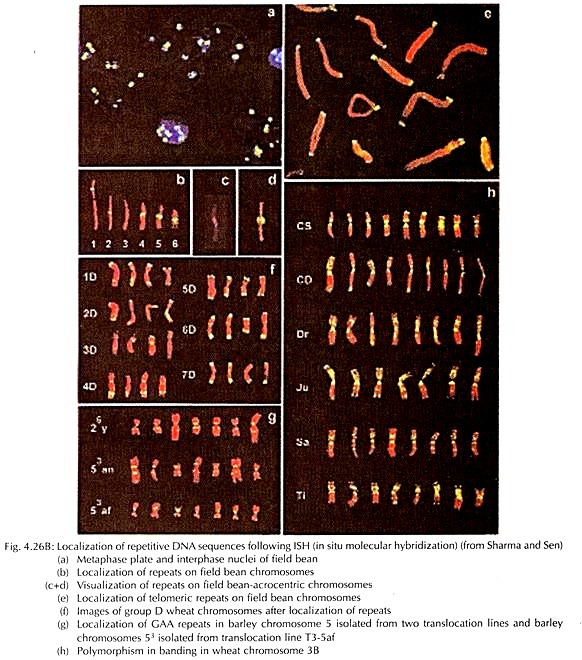
Besides the chromosome banding, for localization and mapping of different segments of chromosomes and gene loci at the microscopic level, application of molecular hybridization in situ is now widely adopted (Fig. 4.26B).
In situ hybridization (ISH) principally uses probe sequences, tagged with radioisotopes or fluorescent compounds (or a chemical reporter). The initial step is denaturation of the target which is followed by hybridization with probe of the complementary sequences to undergo re-annealing or pairing.
The complementary sequences of the probe bind selectively at the target site. The hybridized sites are localized either through autoradiography or immune-fluorescence as well as counter staining with specific stains detected cytologically. The in situ hybridization technique was developed initially by Pardue and Gall and later modified by different authors.
The fluorescent in situ hybridization (FISH) is the most powerful technique at present, through which the target loci at the chromosome is hybridized with complementary probe sequence, tagged with fluorescent compound. There are two approaches, namely direct or indirect, of FISH technique in plant chromosome.
In the indirect method, the probes are tagged with reporter molecule, such as biotin, digoxigenin and finally they are located by fluorochrome conjugated antibodies such as avidin.
The main principle of this method is to make the probe- target sequence as antigenic so that, it can be detected through antibody. The common fluorochromes are FITC (fluorescein-isothiocyanate) as well as rhodamine. In the direct method, the probe is directly labelled by fluorochrome- labelled antibodies. The direct labelling of fluorochrome with the probe is the rapid method of ensuring good resolution.
Due to lack of karyomorphological markers, metaphase chromosome analysis cannot distinguish parental genomes in hybrids. If ISH technique is applied with total genomic probes where the plant has multi-genomic constitution, the parental chromosome can be directly identified in the hybrids, such as Triticum and Secale in Triticale.
Thus method of total genome in situ hybridization is otherwise termed as GISH (genomic in situ hybridization) technique.
Since its first demonstration in identification of parental genomes in hybrid between Hordeum chilense and Secale africanum by Schwarzacher et al., the technique has been extensively applied to elucidate ancestry of hybrids and polyploids. GISH remains a very effective tool in genome identification, their orientation and in establishing genomic relationships between species.
The use of GISH in meiosis helps in understanding inter-genomic homologies as well as in elucidating the possible transfer of chromosome segments through inter-genomic recombination.
The FISH technique, using different colour combinations by different probes, is now being applied to detect simultaneously different genomes, or chromosome segments by extension of the technique – otherwise termed as multi-colour FISH. This method has been used to distinguish three genomes in hexaploid wheat and to detect several translocation sites and insertions in polyploid species of Triticum and Aegilops.
The multi-colour FISH technique is now regarded as a powerful tool for gene mapping as well as detection of abnormalities, including insertion and breakage points with chromosome specific or genome specific dispersed probes. The term chromosome painting is used in FISH technique where chromosome specific dispersed probes are used to detect the location of complementary target sequences in the complement.
The FISH technique in recent years has further been modified to locate single copy or tandem sequences, utilizing primer mediated extension and amplification in situ by PGR method. The application of this method lies also in confirmation of location of foreign gene at the chromosome level in transgenic individuals.