An exclusive project report on Recombinant DNA Technology. This project report will help you to learn about: 1. Introduction to Recombinant DNA Technology 2. Purpose of Recombinant DNA Technology 3. Basic Steps 4. Enzymes Involved 5. Vectors Used.
Contents:
- Project Report on Introduction to Recombinant DNA Technology
- Project Report on the Purpose of Recombinant DNA Technology
- Project Report on the Basic Steps of Recombinant DNA Technology
- Project Report on Enzymes Involved in Recombinant DNA Technology
- Project Report on Vectors Used in Recombinant DNA Technology
Project Report # 1. Introduction to Recombinant DNA Technology:
Genetic engineering involves manipulation of the genetic material towards a desired and in a directed and predetermined way. This method aims at isolating DNA fragments and recombining them; i.e., two DNA molecules are isolated and cut into fragments by one or more specialized enzymes and then the fragments are joined together in a desired combination and restored to a cell for replication and reproduction.
When the recipient organism is a microbe, then as the microbe multiplies, it is possible to obtain millions of copies of a specific region of DNA by allowing the cell to multiply.
Project Report # 2. Purpose of Recombinant DNA Technology:
The interest in genetic engineering principally is due to its varied applications (Fig. 18.1):
i. Production of varieties of plants having particular desirable characteristics (e.g., resistance or tolerance to disease, drought, development of CMS line, etc.).
ii. Improvement in the production of biochemicals and commercially important organic chemicals.
iii. Correction of genetic disorder in higher organisms.
iv. The sequencing of gene, the prerequisite for mapping the genome as well as utilizing the gene for horizontal transfer involved in raising transgenic organism.
Project Report # 3. Basic Steps of Recombinant DNA Technology:
In principle the recombinant DNA technology involves certain basic steps, such as (Fig. 18.2):
i. Isolation of the gene (target DNA) to be cloned.
ii. Insertion of the gene into another piece of DNA called a vector which allows it to be taken up by the recipient cell and replicated.
iii. Transfer of the recombinant vectors into cells of recipient organisms, either by transformation or by infection using viruses.
iv. Selection of those cells which contain the desired recombinant vectors.
v. Growth of the transformed organism.
vi. Expression of the gene to obtain the desired product.
Project Report # 4. Enzymes Involved in Recombinant DNA Technology:
Genetic engineering became possible with the discovery of mainly two types of enzymes: the cutting enzymes called restriction endonucleases and the joining enzymes called ligases.
Restriction endonucleases or restriction enzymes, as they are called popularly, recognize unique base sequence motifs in a DNA strand and cleave the backbone of the molecule at a place within or, at some distance from the recognition site. Whereas ligase is the enzyme that joins a 5′ end of a DNA with a 3′ end of the same or of another strand.
(i) Restriction Endonuclease:
Ordinary nucleases are endonucleases or exonucleases. The former cleaves the DNA backbone between two nucleotides, i.e., it cleaves the double stranded DNA at any point except the ends, but it involves only one strand of the duplex. The latter remove or digest one nucleotide at a time starting from 5′ or 3′ end of a DNA strand.
The restriction endonucleases cleave only at specific regions in a particular DNA, so that discrete and defined fragments are obtained at the end of total digestion. The name ‘restriction’ endonuclease originated from an observation of a system of restriction of the growth of the phage lambda in particular strains of the E. coli host cell.
Most restriction enzymes recognize only one short base sequence in a DNA molecule and make two single strand breaks, one in each strand, generating 3′-OH and 5′-P groups at each position. The sequences recognized by restriction enzymes are often palindromes, i.e., inverted repetition sequences which are symmetrical.
Restriction enzymes can cut DNA in two ways to generate blunt ends (cut precisely at opposite sites, e.g., Hpal) and staggered ends (cut at asymmetrical position, e.g., Eco R!) with short single stranded overhangs at each end. A large number of restriction enzymes have been identified and classified into three categories (type 1, 11, III) on the basis of their site of cleavage.
Restriction enzymes have three important features:
a. Restriction enzymes make breaks in palindromic sequences.
b. The breaks are usually not directly opposite to one another.
c. The enzymes generate DNA fragments with complementary ends.
The commonly employed restriction enzymes are listed in Table 18.1.
(ii) DNA Ligase:
Ends of DNA strands may be joined by the enzyme polynucleotide ligase, called ‘glue’ of the recombinant DNA molecule. The enzyme catalyses the formation of a phosphodiester bond between the 3′-OH and 5′-P terminals of two nucleotides.
The enzyme is thus able to join unrelated DNA, repair nicks in single strand of DNA and join the sugar phosphate backbones of the newly repaired and resident region of a DNA strand.
The enzyme which is extensively used for covalently joining restriction fragments is the ligase from E. coli and that encoded by T4 phage. As the’ main source of DNA ligase is T4 phage, hence, the enzyme is known as T4 DNA ligase.
The ligation reaction is controlled by several factors, such as pH, temperature, concentration and kinds of sticky ends, etc. As ligase uses the, ends of DNA molecules as substrates rather than the entire DNA, the kinetics of joining depend on the number of ends (concentration) available for joining.
(iii) Alkaline Phosphatase:
The broken fragments of plasmids, instead of joining with foreign DNA, join the cohesive end of the same DNA molecules. The treatment with alkaline phosphatase prevents recircularisation of plasmid vector and increases the frequency of production of recombinant DNA molecule.
(iv) DNA Polymerase and the Klenow Fragment:
The DNA polymerase that is generally utilized is either the DNA Pol I from E. coli or the T4 DNA polymerase encoded by the phage gene. The E. coli enzyme is basically a proofreading and repairing enzyme. It is composed of 3 subunits each with a specific activity. They are: 5′-3′ polymerase, 3′-5′ exonuclease and 5′-3′ exonuclease.
The enzyme is useful for synthesizing short length of a DNA strand, especially by the hick translation method. The 5′-3′ exonuclease activity may be deleted, this edited enzyme is referred to as the klenow fragment. The T4 DNA Pol possesses, like the klenow fragment, only the polymerase and proofreading (3-5′ exonuclease) functions.
(v) Reverse Transcriptase:
Retroviruses (possessing RNA) contain RNA dependent DNA polymerase which is called reverse transcriptase. This produces single stranded DNA, which in turn functions as template for complementary long chain of DNA. This enzyme is used to synthesize the copy DNA or complementary DNA (cDNA) by using mRNA as a template.
The enzyme is very useful for the synthesis of cDNA and construction of cDNA clone bank and to make short labelled probes.
Project Report # 5. Vectors Used in Recombinant DNA Technology:
Cloning Vector:
By cloning, one can produce unlimited amounts of any particular fragment of DNA. In principle, the DNA isolated and cut pieces are introduced into a suitable host cell, usually a bacterium such as Escherichia coli, where it is replicated, as the cell grows and divides.
However, replication will only occur if the DNA contains a sequence which is recognized by the cell as an origin of replication. Since such sequences are infrequent, this will rarely be so, and therefore, the DNA to be cloned, has to be attached to a carrier, or vector DNA which does contain an origin of replication.
Criteria of an Ideal Vector:
Vectors are those DNA molecules that can carry a foreign DNA fragment when inserted into it. A vector must possess certain minimum qualifications to be an efficient agent for the transfer, maintenance and amplification of the passenger DNA.
i. The vector should be small and easy to isolate.
ii. They must have one or more origins of replication so that they will stably maintain themselves within host cell.
iii. Vector should have one or more unique restriction sites into which the recombinant DNA can be inserted.
iv. They should have a selectable marker (antibiotic resistance gene) which allows recognition of trans-formants.
v. Vector DNA can be introduced into a cell.
vi. The vector should not be toxic to host cell.
Types of Vector:
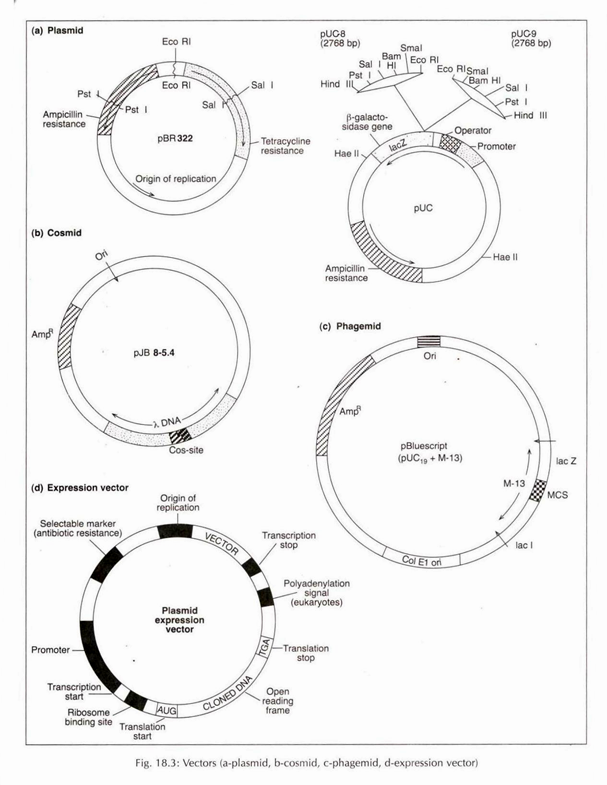
Based on the nature and sources, the vectors are grouped into bacterial plasmids, bacteriophages, cosmids and phagemids (Fig. 18.3).
(a) Plasmid:
Plasmids are the extra-chromosomal, self-replicating, and double stranded closed and circular DNA molecules present in the bacterial cell. A number of properties are specified by plasmids such as antibiotic and heavy metal resistance, nitrogen fixation, pollutant degradation, bacteriocin and toxin production, colicin factors, etc.
Plasmids have following advantages as cloning vehicle:
1. It can be readily isolated from the cells.
2. It possesses a single restriction site for one or more restriction enzymes.
3. Insertion of foreign DNA does not alter the replication properties.
4. It can be reintroduced into cell.
5. Selective marker is present.
6. Trans-formants can be selected easily by using selective medium.
7. Multiple copy numbers are present in a cell.
Some plasmid vectors are pBR 322, pBR 327, pUC vectors, yeast plasmid vector and Ti, Ri plasmids. Ti and Ri Plasmids are widely used in plant system for genetic transformation. Among higher plants, Ti plasmid of Agrobacterium tumefaciens or Ri plasmid of A. rhizogenes are the best known vectors.
T-DNA, from Ti or Ri plasmid of Agrobacterium, is considered to be very potential for foreign gene transfer in cloning experiments with higher plants.
pBR 322 and pUC Vectors:
pBR322 is a derived plasmid from a naturally occurring plasmid col El, composed of 4362 bp DNA and its replication may be more faster. The plasmid has a point of origin of replication (ori), two selectable marker genes conferring resistance to antibiotics, e.g., ampicillin (ampr), tetracycline (tetr) and unique recognition sites for 20 restriction endonucleases.
Tetracycline resistance gene has a cloning site and insertion of foreign segment of DNA will inactivate the tetr gene. The recombinant plasmid will allow the cells to grow only in presence of ampicillin but will not protect them against tetracycline (Fig. 18.3a).
Another plasmid used in gene cloning is pUC vector available in pairs with reverse orders of restriction sites relative to lacz promoter. This is a synthesized plasmid possessing ampicillin resistance gene (ampr), origin of replication from pBR322(ori) and lacz gene from E. coli. pUC 8 and pUC 9 make one such pair (Fig. 18.3a).
(b) Bacteriophage:
The bacteriophage has linear DNA molecule, a single break will generate two fragments, foreign DNA can be inserted to generate chimeric phage particle. But as the capacity of phage head is limited, some segments of phage DNA, not having essential genes, may be removed. This technique has been followed in X (Lambda) phage vectors to clone large foreign particle.
Plasmid can clone up to 20 to 25 kb long fragments of eukaryotic genome. The examples of different Lambda phage vectors are gt 10, X gt 11, EMBL 3, etc. M-13 is a filamentous bacteriophage of E. coli whose single stranded circular DNA has been modified variously to give rise M-13 series of cloning vectors.
(c) Cosmid:
Cosmids are plasmid particles, into which certain specific DNA sequences, namely those for cos sites are inserted which enable the DNA to get packed in λ particle. Like plasmids, the cosmids perpetuate in bacteria without lytic development. The cosmids have high efficiency to produce a complete genomic library (Fig. 18.3b).
(d) Phagemid:
These are prepared artificially by combining features of phages with plasmids. One commonly used phagemid is pBluescript IIKs derived from pUC-19 (Fig. 18.3c).
(e) Plant and Animal Viruses:
A number of plant and animal viruses have also been used as vectors both for introducing foreign genes into cells and for gene .amplification. Cauliflower Mosaic Viruses (CaMV), Tobacco Mosaic Viruses (TMV) and Gemini Virus are three groups of viruses that have been used as vectors for cloning of DNA segments in plant system.
SV 40 (Simian Virus 40), human adenoviruses and retroviruses are potential as vectors for gene transfer into animal cells.
(f) Artificial Chromosomes:
Yeast Artificial Chromosome (YAC) or Bacterial Artificial Chromosome (BAC) vectors allow cloning of several hundred kb pairs which may represent the whole chromosome. It can be cloned in yeast or bacteria by ligating them to vector sequences that allow their propagation as linear artificial chromosome.
(g) Transposons:
Transposable elements like Ac-Ds or Mu-1 of Maize, P-element of Drosophila may also be used for cloning vector and transfer of gene among eukaryotes.
Expression Vector:
A vector that has been constructed in such a way that inserted DNA molecule is put under appropriate promoter and terminator sequences for high level expression through efficient transcription and translation. Example: Use of promoters (‘nos’ from T-DNA) or expression cassettes (pRT plasmids) (Fig. 18.3d).
Shuttle Vector:
There are plasmids capable of propagating and transferring genes between two organisms (e.g., E. coli and A. tumefaciens). It has unique origins of replication for each cell type as well as different selectable markers. It can, therefore, be used to shuttle gene from prokaryotes to eukaryotes.