In this article we will discuss about the procedure for cloning recombinant DNA.
The first step in a gene cloning programme is to construct a recombinant DNA molecule containing a donor DNA segment in which a selected gene is located and a vector DNA. The donor may belong to any taxonomic group. The DNA of the donor is cleaved into fragments using any of the many restriction enzymes.
The vector DNA has also to be cleaved by the same enzyme, so that both DNAs have similar sticky ends. When the donor fragments are mixed with vector fragments, the single-stranded sticky-ends form base-pairs. The free ends are then joined by DNA ligase to obtain a recombinant DNA molecule as shown in Fig. 9.130 and Fig. 9.131 using two different types of vectors.
Although joining of two DNA fragments obtained by digesting them with the same restriction enzyme is not difficult, identification of a recombinant DNA containing a specific donor gene is by no means so simple. Because, the donor DNA is often a very large molecule containing many restriction sites of a single restriction enzyme.
As a result, restriction digest consists of many fragments, only one or a few of these fragments include the gene or parts of the gene to be cloned. The insertion of these fragments into vector yields recombinant DNA, of many sorts, only a few of which contain the desired gene.
In a cloning experiment it becomes essential to identify the particular recombinant molecules in which the gene is present. A straight-forward way of doing this is to identify the gene in a clone by its product. Other methods involve use of a purified DNA containing the gene of choice for insertion into the vector to prepare the recombinant molecule.
The next step is the introduction of the recombinant DNA into a suitable host. This depends on the nature of vector chosen. For insertion of comparatively small DNA fragments, the vector of choice is one of the plasmids which has been suitably tailored for the purpose.
The recombinant circular plasmid vector is introduced into its bacterial host, mostly E. coli, by forced transformation, because E. coli is not normally transformable. The bacteria which have taken up the recombinant plasmid can be identified with suitable markers.
For example, if the plasmid contains a gene conferring resistance to say ampicillin, only those transformed bacteria containing the plasmid can grow in an ampicillin-containing agar and the non-transformed bacteria are eliminated. This is only a preliminary selection. Because of reasons, most of the recombinant plasmids do not have the desired donor gene. For further screening, the technique of colony hybridization is often adopted to identify the bacterial clones with the desired gene.
The cloning procedure using a plasmid vector is shown in a simplified diagrammatic manner in Fig. 9.133:
When bacteriophage vectors are used for cloning, the recombinant DNA obtained by joining the DNA segment of the donor and the processed phage DNA is packaged into the phage head. The recombinant phage particles are allowed to infect appropriate host bacteria by the natural infection process.
The recombinant DNA of the phage multiplies producing large number of progeny phage particles which are released by lysis of the host cells. Recombinant DNA molecules can be isolated from the progeny phage particles for experimental purpose.
The procedure is shown in Fig. 9.134:
The cloning procedure using cosmid vectors is more or less similar to that of phage vectors, except that they are capable of inserting larger DNA fragments of about 40 kb size. Another important distinguishing feature of cosmid vectors is that they multiply in infected hosts as plasmids and do not form phage progeny. Therefore, the question of lysis does not arise.
All the three types of vectors — plasmids, phage and cosmids — are used for cloning recombinant DNA in bacterial hosts, mainly E. coli.
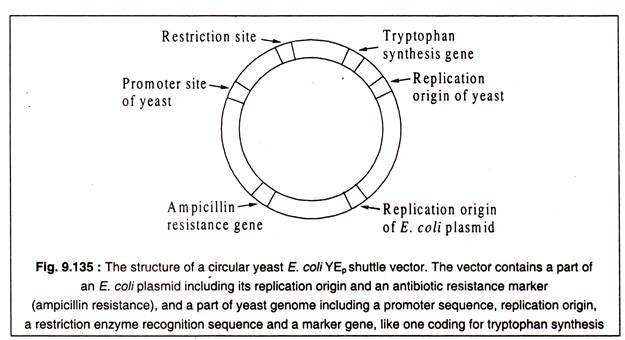
Some- vectors have been developed by genetic engineering techniques which can exist in two different hosts. These are called shuttle vectors. A vector of this type is YEp 24 which can replicate in both yeast and E. coli. These vectors contain sequences of an E. coli plasmid and a part of the yeast genome. Each part has its own replication origin and some other genes as well as restriction sites. A structure of a shuttle vector of yeast and E. coli is shown in Fig. 9.135.
Such shuttle vectors have proved useful in cloning eukaryotic genes like mammalian genes. One problem of cloning eukaryotic genes in prokaryotes is that the bacterial RNA polymerase may fail to initiate transcription of the eukaryotic gene transferred into a bacterial host, because of its inability to interact with the eukaryotic promoter.
Another problem is due to the presence of introns in the eukaryotic primary transcripts (hn-RNA) which the bacteria are unable to remove for they do not have the biochemical machinery. The first problem can be effectively solved by coupling the eukaryotic gene next to a bacterial promoter.
As E. coli is selected generally as the host, its lac-promoter is often used for this purpose. The second problem of removal of introns can be solved, if the eukaryotic gene to be cloned is made free of the intron segments before its joining to the vector DNA.
This can be achieved indirectly by preparing an eukaryotic gene from its m-RNA using reverse transcriptase. The processed form of m-RNA which is translated to yield the gene product is devoid of introns. A complimentary copy of m-RNA produced by reverse transcription gives a DNA without introns. Such a DNA, known as complementary DNA (c-DNA) can then be used for obtaining a recombinant DNA by joining with a suitable vector.