The below mentioned article provides a close view on the Deoxyribonucleic Acid (DNA):- 1. Structure of DNA 2. Alternative DNA Models 3. Single Stranded DNA 4. Localization 5. Different Forms 6. Denaturation and Renaturation 7. Nucleic Acid Hybridization 8. Replication 9. Replication of DNA in Prokaryotes 10. Mechanism of DNA Replication 11. Replication of Viral DNA 12. Replication of DNA in Eukaryotes.
Contents:
- Structure of DNA
- Alternative DNA Models
- Single Stranded DNA
- Localization of DNA in the Cells
- Different Forms of DNA (Conformational Flexibility of DNA)
- Denaturation and Renaturation of DNA
- Nucleic Acid Hybridization
- Replication of DNA
- Replication of DNA in Prokaryotes
- Mechanism of DNA Replication
- Replication of Viral DNA
- Replication of DNA in Eukaryotes
Contents
- 1. Structure of DNA:
- 2. Alternative DNA Models:
- 3. Single Stranded DNA:
- 4. Localization of DNA in the Cells:
- 5. Different Forms of DNA (Conformational Flexibility of DNA):
- 6. Denaturation and Renaturation of DNA:
- 7. Nucleic Acid Hybridization:
- 8. Replication of DNA:
- 10. Replication of DNA in Prokaryotes:
- 11. Mechanism of DNA Replication:
- 12. Replication of Viral DNA:
- 13. Replication of DNA in Eukaryotes:
1. Structure of DNA:
DNA is a polymeric macromolecule consisting of many deoxyribonucleotides in a linear sequence (Fig. 10.3).
Four different deoxyribonucleotides are the major components of DNA which are distinguished by four nitrogenous bases. The four bases characteristic of deoxyribonucleotides are purine derivatives; adenine (A) and guanine (G) and pyrimidine derivatives; thymine (T) and cytosine (C).
In working out the structure of DNA, the following three types of studies have been very helpful:
1. Studies on base pairing and base equivalence:
DNA consists of two unbranched chains of polynucleotides. The backbone of these nucleotide chains is quite regular and consists of alternate sugar and phosphate group joined with 3′ and 5′ phosphodiester linkages. The sugar molecule is attached on one side with a base.
In 1950, E. Chargaff together with his colleagues E. Vischer and S. Zamenhop analysed DNA of several different organisms and pointed out that in DNA the amount of adenine equals the amount of thymine (i.e., A=T) and the amount of guanine equals that of cytosine (i.e., G = C).
This important observation has now become a universal law that holds true for every replicative DNA found in nature. Within the limits of experimental error the ratios of A/T and G/C are equal to 1 except for the isolated coliphage ф x 174. The ratio A+T/G+C is not equal to one and has been used to characterise the DNA from a particular source.
2. Studies based on X-ray diffraction:
While the work on the base composition was proceeding in Chargaff’s laboratory, Rosalind Franklin and Maurice H. F. Wilkins at Kings College London were using x-ray diffraction patterns for the analysis of DNA structure. They observed that DNA from different sources had remarkably similar x-ray diffraction patterns.
Astbury had earlier shown “a 3.4 Å repeat, i.e., the distance between any two consecutive nucleotides in the polynucleotide chain was 3.4 Å. To this Franklin and Wilkins added a 34 Å repeat which was intriguing because nothing in the polynucleotide structure appeared to correspond with this dimension.
With the x-ray diagram. Franklin and Wilkins also showed that DNA molecule is a very long thin helical strand of about 20 Å diameter. Thus, x-ray diffraction study illucidated the geometry of molecules i.e., their arrangement in space and determined the relative position of atoms making up the molecule (Fig. 10.3).
The diffraction pattern obtained by Wilkins and Franklin suggested that the DNA molecule was a double stranded structure. It was in marked contrast to the three stranded structure, Pauling had proposed earlier.
The spatial geometry of DNA was determined to be in the form of an alpha (α) helix, a spiral configuration maintained by intramolecular hydrogen bonds. It was then important to decide whether bases pointed outward or toward each other in the centre of the molecule.
Pauling suggested that bases pointed toward outside but Franklin felt that she had evidence that phosphate pointed toward outside and that the bases were in the centre.
3. Studies based on titration:
The analytical studies suggested that the two long nucleotide chains were held together by hydrogen bonds between base residues. This was the situation in 1951 when James D. Watson, [Fig. 10.3 (a)] a 22-year-old American postdoctoral research fellow, arrived in Cambridge and met Francis H.C. Crick, [Fig. 10 3 (b)], a physicist working for his Ph. D. degree in biophysics.
Although they were to work on different problems, they decided to collaborate on the study of DNA. In 1953, Watson and Crick proposed a double helical model of DNA molecule based on the research of Chargaff, Wilkins, Franklin. Pauling and others.
To explain their model of DNA Watson and Crick postulated several remarkable features which are as under:
1. The DNA molecule is built up of a long chain of deoxyribonucleotides. The chain of nucleotides is referred to as polydeoxyribonucleotide. The sequence of nucleotides in chain makes up the primary structure of DNA (Fig. 10.4). The number of polynucleotide chains in a DNA molecule is two and not three.
2. The chains follow right handed helix with 10 bases in one complete turn of the spiral.
3. The two chains are coiled plectonemically, i.e., in an interlocked way about the same axis (Secondary structure).
4. Polynucleotide chains have polarity. The pentose sugar at one end of the chain has 5′ hydroxyl or phosphoryl group (5′ end) and the sugar at the other end has a 3′ hydroxyl group (3′ end) as shown in Figure. One of the chains ascends and the other descends (i.e., two chains have reversed polarity or in other words sequences of the atoms in two chains run in opposite directions) (Figs. 10.5 & 10.6).
Thus if one strand of polynucleotides has phosphodiester linkages established in 3’—>5′ direction, the complementary strand has phosphodiester bonds in just reverse or 5′<—3′ direction. In natural polynucleotides, the entire chain has polarity, i.e., all the 5′ carbons point in the same direction so that the chain must end in 5C at one end and 3C at the other end.
5. In the double helix the phosphates of the nucleotides are on the outside and the bases on the inside. The nucleotides are set in the planes at right angles to the axis of the helix and spaced at intervals of 3.4 Å or 0.34 nm. (1 nm =1 nanometer = 10-9 m).
6. The two chains are held by hydrogen bonds which are established between the base pairs, or in other words bases from one polynucleotide chain are hydrogen bonded to bases of the opposite chain.
7. The pairing is highly specific because there is a fixed distance of 10 or 11 A(1.11 nm) between the two sugar moieties in the opposite nucleotides. Purine base normally pairs with a pyrimidine base (Fig. 10.3). Thus A—T, C—G, T—A and G—C pairs are formed. A and T are joined with 2 hydrogen bonds and C and G are joined with three hydrogen bonds (Figs. 10.6 and 10.7).
Adenine cannot pair with cytosine because there would be two hydrogen atoms near one of the bonding positions and none at the other. Similarly guanine cannot pair with thymine.
There is no H-bonding between A and C or G and T.
It is true that hydrogen can form only one true covalent bond but under certain conditions hydrogen which is chemically linked to one atom can form a weak linkage of non-covalent type to a second atom also, i.e., covalent bonded hydrogen atom with its partial positive charge is shared between two covalently bonded nitrogen atoms or an oxygen atom and a nitrogen atom, both with partially negative charges.
Hydrogen bonds are much weaker than ordinary covalent bonds. It follows from the Watson and Crick model of DNA that the base sequences of the two strands are complementary. The sequence of bases in a polynucleotide chain is not definite.
The base sequence is fixed for a particular DNA strand and it varies from strand to strand.
Thus the sequence of bases in a segment of hypothetical DNA molecule might be as follows:
Watson and Crick model found immediate support from the works of Wilkins, Stokes and Wilson (1953), Franklin and Gosling (1953), Feughalman, Langridge, Seeds, Hooper, Hemilton et al. (1955). For this brilliant piece of work Watson, Crick and Wilkins were jointly awarded Nobel Prize in 1962.
Actually, this work was jointly conducted by many scientists. Dr. Watson said sometime later, “actually it was matter of 5 persons: Maurice Wilkins, Rosalind Franklin, Linus Pauling, F. H. C. Crick and me.”
In the cases where DNA double helix does not exist with free ends, the DNA molecule is freely movable and can take over any shape. In some cases the molecule becomes a close ring. Such twisted DNA double helix is said to have a tertiary structure, as for example, DNA in bacteria, plastids, mitochondria and some viruses.
2. Alternative DNA Models:
The double helix model for DNA structure proposed by Watson and Crick has been the focal point in biology for more than 20 years. The impact and importance of the model has been so great that scientists did not feel any necessity to re-examine the x-ray diffraction data on DNA structure proposed by Watson & Crick.
It is only recently that physical scientists have begun to re-examine x-ray data interpreted by Watson and Crick and suggest some alternative conformations for DNA.
It is established beyond doubt that DNA consists of two polynucleotide chains held together by hydrogen bonds between specifically paired bases. But the way in which the polynucleotide chains are associated has been challenged in one particularly alternative structure.
In this alternative structure, DNA are viewed as side by side double helix rather than being interlocked double helix as suggested by Watson & Crick. G. A. Rodley et al. (1976) working in New Zealand and V. Chandrasekharan et al. in India have independently proposed a structure of B DNA which is different from the model suggested by Watson and Crick.
According to them DNA is formed of alternative right handed and left handed helices arranged side by side. This structure has been called right-left handed helix (RL helix) or side by side double helix.
Although interlocked and side by side or RL versions of double helix are compatible with x-ray scattering and other physical data, yet side-by-side model does have one interesting feature from the genetic point of view. There has always been some doubts as to how the strand separation and unwinding of the Watson & Crick’s interlocked double helix could be achieved during DNA replication.
The RL helix makes such separation much easier to achieve than the Watson and Crick model because in the latter case a moment of force or torque would develop in DNA strands during unwinding and mere breaking of hydrogen bonds between two strands would make them free. The RL model can also easily explain the super-coiling of DNA in eukaryotic chromosomes and its replication.
3. Single Stranded DNA:
DNA molecule in most organisms consists of two polynucleotide chains that are wound around each other to form a double helix. But in bacteriophage virus ф x 174 and several other bacteriophages, DNA has been found to be single stranded.
Single stranded DNA differs from double stranded DNA in the following respects:
1. Double stranded DNA shows constant absorption of ultraviolet rays from 0 to 80°C (80°C being the critical melting point) and beyond 80°C absorption rate rises rapidly. The single stranded DNA shows steady increase in absorption of UV rays from 20°C to 90°C.
2. Double stranded DNA is resistant to the action of formaldehyde whereas the single stranded DNA is not resistant because the reactive sites are exposed.
3. In double stranded DNA amount of adenine is equal to thymine and G is equal to C. But in single stranded DNA of ф x 174 amounts of A and G are not equal to those of T and C respectively and the proportion is 1: 1.33 and 0.98: 0.75.
4. Single stranded DNA is circular while the double stranded is linear.
During replication single stranded DNA becomes double stranded (replicative form).
4. Localization of DNA in the Cells:
DNA can be located in the cells in the following ways:
1. Feulgen staining:
Feulgen (1912) found that when DNA is hydrolysed with warm Schifif’s reagent it turns reddish purple. During this reaction Schiff’s reagent reacts with aldehyde groups in sugar deoxyribose. In 1924 Feulgen developed basic fuchsin staining technique which gives positive indication of presence of DNA in the chromosomes.
2. Deoxyribonuclease reaction:
DNA can be removed by a specific enzyme deoxyribonuclease. After removal of DNA, the nucleus does not give the Feulgen reaction.
3. DNA absorbs ultraviolet rays at wave-length of 2600 A. By this method it is possible to locate DNA without staining the chromosomes. Caspersson and others have used this technique to measure nucleic acid contents of the nuclei.
5. Different Forms of DNA (Conformational Flexibility of DNA):
DNA can exist in five different forms; A, B, C, D and Z forms of which B form (B DNA) is the most common of the configurations. B DNA is a right handed double helix in which there are 10.0 base pairs per turn (34 Å) of the helix and base pairs are more or less perpendicular to the helix axis.
The structure of DNA molecules changes as a function of their environment. The exact conformation of any DNA molecule or segment of DNA molecule will depend on the interaction between molecule and its environment. When humidity is high, it exists in B-form.
When humidity is relatively low DNA exists in A-form which has 11 base pairs each turn. Since A-DNA occurs under low humidity conditions it has been suggested that it may be formed physiologically as a result of interaction with hydrophobic molecules or changing cellular conditions.
Other forms i.e., C-DNA, D-DNA, and Z-DNA differ from B-DNA in respect of the direction of helical coiling and/or spacing and inclination of base pairs (Fig. 10.8).
In 1979, Andrew Wang, Alexander Rich and their associates of MIT, U.S.A. have proposed a new model of DNA called Z-DNA (named because of zig-zag appearance of the sugar-phosphate backbone). Z-DNA is a left handed double helix in which the base pairs are arranged peripherally to the helix axis and there are 12.0 base pairs per turn of the helix.
B and Z forms of DNA are inter-convertible and are the major forms of natural DNA.
The other two forms, C-DNA and D-DNA have been described both in natural and synthetic states. They are right handed double helical forms which have 7.9-9.6, and 8.0 pairs of bases per turn respectively. In A-DNA the base pairs are considerably tilted from the axis of helix at about 19° angle.
Because of this displacement, the depth of the deep groove is increased and that of the shallow groove decreased, axial rise being only 2.56 Å.
In C-DNA, the base pairs are arranged towards the middle of the helix and are inclined at an angle of 7.8° (less than that of A-DNA). D-DNA, as viewed along the helix axis, is not circular but hexagonal in cross section. In this form the base pairs are arranged towards the middle of the helix and are tilted at an angle of 16.7° from the axis of the helix.
6. Denaturation and Renaturation of DNA:
Soon after the publication of Watson and Crick’s model of DNA, many experiments were carried out by Warner, Rich and others using synthetic polynucleotide strands which showed that the complementary strands have marked tendency to intertwine in pairs and form right handed double helices.
Recent investigations of DNA in solution or when packed into native virus particles have shown that the double helix often departs from perfect regularity and may partially untwist to permit local separation of two polynucleotide chains (called “breathing”). The two polynucleotide strands of a DNA molecule remain held by weak hydrogen linkages. Thus they can easily be separated.
R.B. Inman (1966) showed that if A. DNA is treated with alkali (high pH) for short periods, DNA strands in the region rich in A—T pairs tend to separate before regions containing more G—C. This is called partial denaturation or partial melting.
The reason of this partial denaturation is that an A—T pair shares two hydrogen bonds, whereas a G—C pair shares three hydrogen bonds. By preventing the complimentary strands from further pairing through the use of formaldehyde, Inman observed a highly reproducible pattern of “bubbles” along the length of DNA molecule.
This pattern is called denaturation map. Paul Doty and Julius Marmur (1960) observed that when DNA is heated to 100 °C or slightly below the boiling point of water, all the hydrogen bonds between the complementary bases are destroyed and the DNA becomes single stranded. This is called melting or denaturation.
If the solution is cooled slowly, some double stranded DNA may result which may be normal. This process is called renaturation or reannealing. The renaturation occurs when two single strands run parallel to each other in such a way that the complementary base sequences align.
This makes, it possible for different DNAs to reanneal after denaturation and mixing. So, annealing provides a measure of similarity between two different DNAs. The property of renaturation has been used to recognize which segment of DNA has been used as a templet for a given RNA species.
Renaturation studies have led to the discovery of repeated sequences of DNA in eukaryotic cells. When certain base sequences are repeated many times the rate of renaturation will be faster than the sequence represented only once.
7. Nucleic Acid Hybridization:
Reannealing or renaturation of DNA is an important tool in molecular biology. It allows inter molecular hybridization between two single stranded DNAs of different species. This is called DNA/DNA hybridization. Since the hybridization involves parts of DNA strands which have complementary base sequences, the amount of hybridization will indicate the degree of genetic similarity.
This technique has been developed by M.Pardne and J. Gall (1970). By this technique single strand of DNA may be hybridized with RNA by complementary base pairing.
In nucleic acid hybridization experiment DNA double helices are heated so that they denature into single strands: At this stage the denatured DNA strands are mixed with single-strand fragments of DNA or RNA obtained from other source (i.e., another plant or animal species).
The mixture is then allowed to cool slowly so that reannealing may take place. If the fragments and the denatured DNA carry homologous base sequences, hybrid DNA/DNA or DNA/RNA duplexes will be formed. Hybridization will fail if homologous base sequences are not available in the fragments involved.
8. Replication of DNA:
There are three possible methods of replication of double stranded DNA, namely:
(i) Conservative,
(ii) Dispersive, and
(iii) Semi-conservative.
(i) Conservative DNA replication:
In this mechanism of DNA replication two parental polynucleotide chains of DNA remain together and the newly formed polynucleotide chains form the daughter molecule of DNA.
(ii) Dispersive DNA replication:
In this replication mechanism, the two helical strands are broken along their length and produce small fragments. Each segment of the broken strands replicates and then all become randomly connected to form two new molecules.
(iii) Semi-conservative replication:
According to Delbruck and Stent (1951), the two strands of a DNA molecule would separate from each other, maintain their integrity and both the strands will synthesize from the pool of nucleotides their complementary strands. The result would be that the newly synthesized molecules would carry or conserve one of the two strands of parent molecule.
In this process the synthesis of DNA strands goes hand in hand with replication and is not complete till the synthesis of new polynucleotide chains.
The greatest attraction of the Watson and Crick DNA model for the geneticists is its ability to provide a simple mechanism for gene duplication. These two workers suggested that the two strands of a DNA molecule manufacture the replica of their own and in the process of replication each DNA strand might serve as a template (base) for the synthesis of its complementary strand (semi-conservative method).
Watson and Crick hypothesized that:
1. Before duplication, the H-bonds are broken. Then unwinding and separation of the two chains take place.
2. Then follows the formation of new complementary polynucleotide chains along the sides of old DNA strands. So two pairs of polynucleotide chains are formed from the original one pair. In the semi conservative model of replication, each newly formed double helix of DNA contains one old and one new polynucleotide strand.
According to the present day theory, the duplication of DNA in living cells including bacteria, plants and animals takes place in semi-conservative manner (Fig. 10.9).
As suggested by Watson and Crick, the two strands uncoil and separate. Once the two strands are free, the base sequences which according to present view carry the genetical codes are exposed to the surrounding nuclear fluid that contains all kinds of chemical building blocks.
The DNA molecule is assembled from precursor deoxyribonucleotide 5-triphosphates namely deoxyadenosine triphosphate (d-ATP),deoxyguanosine triphosphate (d-GTP), thymine triphosphate (TTP) and deoxycytidine triphosphate (d-CTP) in presence of the enzyme DNA-polymerase (Fig. 10.10).
3. The bases of separated DNA strands now attract their complementary bases towards them. Each base in the strand would attract in correct position the particular base that is its partner (i.e., A chooses T, T attracts A, G chooses C and C chooses G). These bases reach to the DNA strands in the form of their respective nucleoside triphosphates.
Nucleoside mono-phosphates and diphosphates are not directly incorporated in DNA.
The cellular enzymes that catalyse the synthesis of nucleic acid can act only on the nucleoside triphosphates and remove two of the three phosphates of each nucleoside triphosphate releasing a tremendous amount of energy and involving the third phosphate group in phosphodiester bond.
Since the two strands of old DNA molecule have reverse polarity i.e., one strand has phosphodiester bonds in 3’—5′ direction and the other has phosphodiester linkages in just reverse or 5’—3′ direction, the phosphodiester bonds in the new strands will be established in the directions just reverse to those of old strands (Figs. 10.11, 10.12).
The newly synthesized chain starts with a 5′ phosphate end. Two terminal inorganic phosphates of deoxyribonucleoside triphosphate are split off as pyrophosphates and a phosphodiester bond is established between the 3′ OH group of a nucleotide with 5′ phosphate group of the next nucleotide in the newly developing DNA strand in 5′ —3′ direction (Fig. 10.13).
Watson and Crick theory of DNA replication was tested by Messelson and Stahl in 1958. Using the isotopic and centrifugation techniques they confirmed the mechanism of DNA replication and called that type of replication as semi-conservative replication because each of the two resultant daughter DNA molecules retains or conserves one parental polynucleotide strand (Fig. 10.14).
The synthesis of DNA in vitro (i.e., in test tube) has been demonstrated by Kornberg and his associates in 1958. Lehman, Bessman, Sims and Kornberg (1958) purified an enzyme from the bacterium Escherichia coli.
They took DNA subunits (different types of deoxyribonucleoside triphosphates), specific enzyme-DNA-polymerase and the isolated DNA strands to demonstrate the DNA synthesis in virto. DNA strand was a necessary primer.
The DNA strands obtained from different sources can be used as primers in the resynthesis.
In the experiment they demonstrated a newly made DNA which was complementary to primer DNA. Bessman, Lehman, Sims and Kornberg (1958) have shown that in the process of polymerisation, the triphosphates retards the rate of reaction and they have also shown that no synthesis of new DNA took place in the absence of primer DNA.
The synthesis of DNA in test tube strongly supports template hypothesis of DNA replication as proposed by Watson and Crick.
This has been fully confirmed again by investigations of Professor H.G. Khorana and his associates at the Institute of Enzyme Research in the University of Wisconsin. It is possible to make short polynucleotide chains analogous to DNA by purely chemical processes.
Thus d A (Deoxyadenylic acid) and d C (Deoxyribocytidylic acid) were induced to combine to form a dinucleotide, i.e., d AC and then six of these dinucleotides were combined to give the repeating unit d AC AC AC AC AC AC.
In the same way a stretch of nucleotides containing complementary unit TG was put together. On mixing these two products they obtained, a small piece of double stranded DNA containing alternately arranged A and C in one strand and T and G in the other. When that DNA was mixed with the DNA polymerase and the necessary precursors, it was found that extensive DNA synthesis occurred.
10. Replication of DNA in Prokaryotes:
Meselson and Stahl experimentally demonstrated that the replication of circular DNA in E. Coli u semiconservative type but they did not explain whether the replication is initiated at one point or at several points simultaneously.
Cairns (1963) provided conclusive evidence on the basis of visual observation of replicating chromosome in which DNA was labelled with tritiated thymidine (3H TdR) that the bacterium E. coli contained single initiation point for the replication of circular DNA which proceeds bidirectionally from that point.
The initiation point (ori) is a unique gene region from where the synthesis of new DNA proceeds outwardly in both clockwise and anticlockwise directions until both ends meet on the opposite side of DNA circle.
Now at least three different DNA polymerases with almost similar catalytic properties have been distinguished in E. coli.
These DNA polymerases are:
(i) DNA polymerase I (DNA pol I),
(ii) DNA polymerase II (DNA pol II), and
(iii) DNA polymerase
(III) (DNA pol III).
It appears that DNA polymerase III is the main enzyme which is concerned with the DNA replication.
Although DNA pol I also takes part in DNA replication, it is largely used in DNA repair process, i.e., for filling the gaps between small Okazaki fragments that are formed during DNA synthesis.
As regards DNA poI II, no specific function has been attributed to that.
DNA pol l which is a single polypeptide chain with 1,000 aminoacid residues has a mol. wt. of about 1,09,000 catalyses (i) the addition of deoxyribonucleotide units to the free 3′ OH end of a polypeptide chain, (ii) catalyses 3′ – 5′ exonuclease activity, and (iii) catalyses 5′ – 3’exonulease action involving hydrolysis of mononucleotide residues from 5′ terminal. The exonuclease activity functions like a correcting type writer so that incorrect base pairs are detected and cut to allow accurate replication. This is called proofreading and it is responsible for high degree of precision in DNA Synthesis.
DNA pol II enzyme has mol. wt of about 1,20,000 and it catalyses 3′ – 5′ exonuclease activity. It is unable to replicate long single strand with a short complementary primer.
DNA pol III is the most active enzyme among the three DNA polymerases. Biologically active form of DNA pol-III* requires an auxilliary protein called DNA copolymerase III* (DNA Copol III*). Active DNA pol-III* makes complex with Copol III*.
The DNA pol III* – Copol III* complex (Fig. 10.15), DNA template strands, RNA primer and ATP are required to initiate replication of DNA. After the initiation of DNA replication, Copol III becomes free from the complex and ADP and inorganic phosphate (Pi) are released.
11. Mechanism of DNA Replication:
As far as the initiation of replication is concerned the process involves the following steps:
(i) Recognition of origin of initiation point.
(ii) Unwinding of double helix and separation of two DNA strands.
(iii) RNA priming.
(iv) Continuous synthesis of a new leading strand on one DNA template in 5′-3’direction and formation of small DNA fragments beyond RNA primers on the second DNA template strand to produce new lagging strand.
i. Recognition of origin of initiation point:
None of the three DNA polymerases known in prokaryotes and eukaryotes can initiate new DNA chains. Instead, initiation of DNA synthesis involves synthesis of a short primer to which deoxyribonucleotides are added by action of DNA polymerases.
Before DNA polymerase starts its action, presumably at several points in duplex DNA, some unique proteins of low molecular weights bind to one of the strands especially in those regions which are rich in A = T base pairs and are susceptible to separation of the two strands of DNA.
ii. RNA priming:
DNA replication is discontinuous and always proceeds in 5′-3’direction on both strands of duplex DNA [Fig. 10.15 (a)]. The next step, after recognition of initiation point is the generation of short RNA primers, i. e., fragments of RNA which develop from precursors ribonucleoside triphosphate by the action of RNA polymerase or primase.
The primer base sequence is directed by the nucleotide sequence of DNA template strand and are 50-100 nucleotides long.
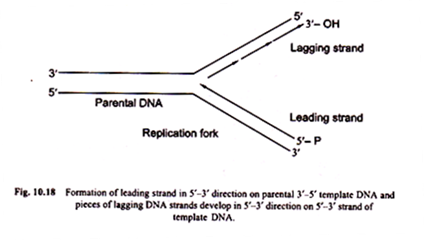
Initiation of replication process starts by RNA polymerase which recognises the initiation point (Fig. 10.16). After formation of RNA primer, DNA polymerase III begins to add deoxyribonucleotide units that are generated from deoxyribonucleotide triphosphates to 3′ terminal of RNA primers. These R-DNA fragments are called Okazaki fragments. They grow in 5′-3’direction (Fig. 10.17).
iii. The leading versus lagging strands:
There is a difference in the types and the rates of growth of two daughter strands formed on two DNA templates. The strand formed on the old 5′ – 3′ DNA template is called the leading strand. The leading strand grows in 5′ – 3′ direction continuously and moves slightly ahead of the other strand.
The second new strand develops on old 3′ – 5′ template strand grows in direction opposite to the direction of movement of the replicating fork.
The strand developing on 3′ – 5′ old strand grows discontinuously and is assembled from a group of DNA fragments made individually in 5′-3’direction. That is called lagging strand (Fig. 10.18).
Thus, following the initiation a number of short segments of DNA are formed in presence of DNA polymerase and ATP.
During the process ATP is hydrolysed to ADP and Pi. and as soon as the elongation process is completed the following three important events occur (Fig. 10.19):
(i) Excision of RNA primer fragments [Fig. 10.19 (a)].
(ii) Excision of RNA primers is followed by filling up of the gaps by deoxyribonucleotide residues [Fig. 10.19 (b)].
(iii) Joining together of adjacent ends of DNA fragments by phosphodiester bonds catalysed by DNA Ligases to form a continuous DNA daughter strand [Fig. 10.19 (c)].
Separation of RNA primer takes place in presence of specific enzyme protein. It is possible that the RNA primer is excised residue by residue from 5′ end by exonuclease activity of DNA polymerase I whose primary task is to fill up the gaps left between RNA primed DNA segments (Okazaki segments). Finally adjacent 5′ and 3′ ends are joined by DNA ligase or polynucleortide ligase by formation of phosphodiester bonds.
iv. Replication of bacterial DNA:
DNA thread of bacterial cell is regarded as bacterial chromosome or genophore or chromoneme. In the case of Esherichia coli the DNA strand contains about 3 million pairs of nucleotide bases and this is sufficient to form nearly 10,000 genes.
The total time taken for replication of DNA in Escherichia coli is 40 to 50 minutes at 37°C which means that the synthesis of DNA takes place at the rate of about 1,000 base pairs per second—a very rapid process if done by only one enzyme molecule.
Replication of DNA is brought about, as already mentioned, by the enzyme DNA polymerase and the replication starts at the one point of circular chromosome and goes round the circle until a new circle is completed, subsequently, the two daughter chromosomes can be separated.
It was shown by Francois Jacob, Sidney Brenner and F. Cuzin (1963) that the replication in fact occurs at the point at which the chromosome is attached to the plasma membrane. Here the replication enzyme DNA polymerase is located and it appears that instead of enzyme moving round the DNA ring, the latter (DNA) moves through the point of attachment called swivel point and is replicated (Fig. 10.20).
12. Replication of Viral DNA:
Viruses contain a nucleic acid and protein sheath. Nucleic acid is DNA in some and RNA in others. Viral nucleic acid contains all the genetic information necessary for the construction of new viruses.
In almost all the viruses genetic material appears to be a single DNA or RNA molecule, either single or double stranded, ranging in length for 1.8 µ in φ x 174 to 56 µ in T2 virus. DNA of T4 phage virus is circular and has a molecular weight 1.3x 1010 (i.e., 2,00,000 nucleotide pairs) which is sufficient for 150 to 200 genes. DNA of T2 phage has about 60,000 base pairs. Coli phage lambda virus has 32 to 50 genes.
Bacterial virus φ x 174 contains single ring shaped DNA strand containing about 5,500 nucleotides (mol. weight 1.7 x 106). The replication process of φ x 174 phage virus is very remarkable and extraordinary series of changes takes place when viral nucleic acid is injected into bacterial cell.
The normal functions of the cell, e.g., RNA and protein syntheses are disturbed. In the host cell the viral DNA is immediately replicated and as a result double stranded form is obtained. The original strand is called positive (+ strand) and the newly synthesized the negative strand (- strand). The two remain associated and form a covalently closed superhelical structure known as parental replicative form (R.F.).
The double stranded R.F. is capable of carrying the usual function of DNA, i.e., it produces messenger RNA which become attached to the ribosomes of host cell to make a whole series of new viral proteins and enzyme required for the replication of phage DNA.
It has been found that the special protein which is produced by the virus in the early stages of infection checks the synthesis of host DNA. Besides acting as template for forming messenger RNA, the DNA must also arrange for its own replication. This is brought about by a special DNA polymerase enzyme which is formed within the cell at an early stage after infection.
The parental R.F. undergoes several rounds of semiconservative replication to produce perhaps some 15 double stranded daughter R.F. molecule. Replication of DNA can occur only at one site within the cell at any time.
This site is probably the site of the cell membrane at which replication of bacterial DNA strand normally occurs. Eventually, a shift occurs and positive stranded rings rather that RF duplexes are synthesized. The rings are packed into virus particles and lysis of the host cell occurs.
There are phages which contain RNA instead of the mere usual DNA. The remarkable feature of RNA of these viruses is that it is capable of acting both as a template for replication and also as mRNA for protein synthesis.
The RNA in the virus is in single stranded form and when RNA enters the cell it first acts as messenger for protein synthesis and then it is converted into double stranded form which is capable of replication through the action of special replicative enzyme.
13. Replication of DNA in Eukaryotes:
The duplication of nuclear DNA takes place during S-phase of cell cycle. J.H. Tailor in 1957 showed that DNA replication in prokaryotes is semi-conservative and DNA replication in eukaryotes is semidiscontinuous. Like prokaryotes, the eukaryotic cells also possess DNA polymerases and DNA ligases.
In eukaryotes three DNA polymerases; α(alpha), β (beta) and Y (gamma) DNA polymerases have been distinguished on the basis of molecular weight, chromatographic properties, sensitivity to N-ethylmaleimide, salt effect and template copying property. The characteristics of these polymerases have been compared in Table 10.3.
Some eukaryotes have multiple forms of DNA polymerases and some others have single major enzyme, a and 3 DNA polymerases along with some other proteins usually occur in the replication complex of nuclei and Y DNA polymerases are localised in the mitochondria. Protozoa, fungi and plants do not have P polymerases.
The eukaryotic microbes, in general, have a polymerase which is different from a DNA polymerase of mammals. There is too much variability among these DNA polymerases and so it is not possible to make generalization about these enzymes, a DNA polymerase is concerned with the nuclear DNA replication and P polymerase probably functions as a repair enzyme.
Eukaryotic DNA ligases require ATP instead of NAD as adenyl donor.
As regards the initiation of DNA replication. Cairns (1963) on the basis of visual observation of replicating chromosome of E. Coli in which DNA was labelled with tritiated Thymidine (3H-TdR) pointed out that in bacterium, the replication of circular DNA is initiated at one point. In eukaryotic organisms replication of DNA begins at several precisely defined units along the double stands of DNA.
These units are called replication units (RUs) or replicons. Each replication unit has a single specific origin (o) and two termini (t) (Fig. 10.21). In Hela cells (a type of tissue culture cells derived from human cancer cells) the chromosomes may contain as many as 100 such RUs each.
This arrangement of RUs in chromosomes is necessary so that enormous length of eukaryotic DNA can be replicated within reasonable time. The spacing between origin points of two adjacent RUs varies from 30,000 to 3,00,000 base pairs.
DNA replication in eukaryotic chromosomes begins by opening of bubbles. The area where double stranded DNA molecule becomes untwisted or denatured for replication is called replication bubble. The breakage of H bonds holding the complementary base pairs together is followed by unwinding or untwisting of two DNA strands results in separation of the DNA chains in the replication units.
The two separated single strands of DNA act as templates on which new complementary strands of DNA will develop. One or more specific initiation proteins are required to recognise these initiation or origin points. In linear DNA molecules, replication starts on specific initiation points internally and generates new complementary strands which grow bidirectionally towards termini.
Though origin and termination signals are little known, perhaps some specific nucleotide sequences act as replication sites which signal the initiation of DNA replication in certain animal viruses such as SV-40 and yeasts.
At replication point, specific initiation protein of replication complex binds to a genetically determined origin sequence to start DNA replication. The rate of movement of replication forks may range from 1,000 to 15,000 nucleotides of DNA per minute at 37°C in E. coli.
In eukaryotes the replicating fork moves slowly at about 2,600 bases per minute. In Drosophila egg cell about 6,000 replicating units are involved in replication and the replication is completed in about 3 minutes.
In eukaryotic chromosomes initiation of replication of each replication unit begins at origin of replication (o), proceeds bidirectionally producing replication bubbles in DNA and the process terminates after fusion of adjacent replication forks. G-C rich regions of DNA are replicated first and A-T rich regions later during S phase.
DNA synthesis also depends on RNA synthesis but the inhibition of RNA synthesis does not have any immediate effect on DNA synthesis. Genetic and biochemical evidences suggest that the start of S phase of cell cycle is determined by a regulatory protein synthesised during GI phase. That protein acts as positive effector to derepress DNA replication.
The newly formed DNA in eukaryotes becomes complexed with histones to form nucleosomes.
The distribution of old and newly synthesised histone octomers in replicated DNA is not clearly understood but there is evidence that there is no mixing of old and new histone octomers in nucleosomes and the old octomers are conserved from generation to generation and the newly synthesised histone octomers are used at the replication forks to form new nucleosomes.
Once the nucleosomes are assembled at the replication forks, within a short period, say 15 minutes, the chromatin matures into non-replicating state.