Read this article to learn about the history, types, structure, silent features and functions of DNA:
Contents
Historical:
Nucleic acids were first isolated by Friedrich Miescher (1869) from pus cells.
They were named nuclein. Hertwig (1884) proposed nuclein to be the carrier of hereditary traits. Because of their acidic nature they were named nucleinic acids and then nucleic acids (Altmann, 1899).
Fisher (1880s) discovered the presence of purine and pyrimidine bases in nucleic acids. Levene (1910) found deoxyribose nucleic acid to contain phosphoric acid as well as deoxyribose sugar.
He characterised four types of nucleotides present in DNA. In 1950, Chargaff found that purine and pyrimidine content of DNA was equal. By this time W.T. Astbury had found through X-ray diffraction that DNA is a polynucleotide with nucleotides arranged perpendicular to the long axis of the molecule and separated from one another by a distance of 0.34 nm.
In 1953, Wilkins and Franklin got very fine X-ray photographs of DNA. The photographs showed that DNA was a helix with a width of 2.0 nm. One turn of the helix was 3.4 nm with 10 layers of bases stacked in it. Watson and Crick (1953) worked out the first correct double helix model from the X-ray photographs of Wilkins and Franklin. Wilkins, Watson and Crick were awarded Nobel Prize for the same in 1962.
Watson and Crick (1953) built a 3D, molecular model of DNA that satisfied all the details obtained from X-ray photographs. They proposed that DNA consisted of a double helix with two chains having sugar phosphate on the outside and nitrogen bases on the inner side.
The nitrogen bases of the two chains formed complementary pairs with purine of one and pyrimidine of the other held together by hydrogen bonds (A-T, C-G). Complementary base pairing between the two polynucleotide chains is considered to be hall mark of their proposition. It is of course based on early finding of Chargaff that A = T and С = G Their second big proposal was that the two chains are antiparallel with 5’→ 3′ orientation of one and У → 5’orientation of the other.
The two chains are twisted helically just as a rope ladder with rigid steps twisted into a spiral. Each turn of the spiral contains 10 nucleotides. This double helix or duplex model of DNA with antiparallel polynucleotide chains having complementary bases has an implicit mechanism of its replication and copying.
Here both the polynucleotide chains function as templates forming two double helices, each with one parent chain and one new but complementary strand. The phenomenon is called semi conservative replication. In vitro synthesis of DNA has been carried out by Kornberg in 1959.
Types of DNA:
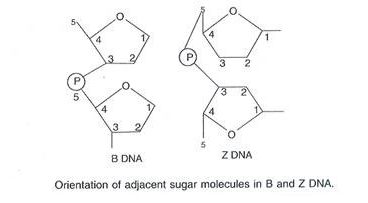
DNA duplex model proposed by Watson and Crick is right handed spiral and is called B-DNA (Balanced DNA). In the model the base pairs lie at nearly right angles to the axis of helix (Fig. 6.5 D). Another right handed duplex model is A-DNA (Alternate DNA). Here, a single turn of helix has 11 base pairs.
The base pairs lie 20° away from perpendicular to the axis. C-DNA has 9 base pairs per turn of spiral while in D-DNA the number is only 8 base pairs. Both are right handed. Z-DNA (Zigzag DNA) is left-handed double helix with zigzag back-bone, alternate purine and pyrimidine bases, single turn of 45 A length with 12 base pairs and a single groove.
B-DNA is more hydrated and most frequently found DNA in living cells. It is physiologically and biologically active form. However, it can get changed into other forms. Right handed DNA is known to change temporarily into the left handed form at least for a short distance. Such changes may cause changes in gene expression.
Circular and Linear DNA:
In many prokaryotes the two ends of a DNA duplex are covalently linked to form circular DNA. Circular DNA is naked, that is, without association with histone proteins, though polyamines do occur. In linear DNA the two ends are free. It is found in eukaryotic nuclei where it is associated with histone proteins.
Linear DNA, without association with histone proteins, also occurs in some prokaryotes, e.g., Mycoplasma. In semi-autonomous cell organelles (mitochondria, plastids) DNA is circular, less commonly linear. It is always naked.
Chargaff’s Rules:
Chargaff (1950) made observations on the bases and other components of DNA. These observations or generalizations are called Chargaff’s base equivalence rule.
(i) Purine and pyrimidine base pairs are in equal amount, that is, adenine + guanine = thymine + cytosine. [A + G] = [T + C], i.e., [A+G] / [T+C] = 1
(ii) Molar amount of adenine is always equal to the molar amount of thymine. Similarly, molar concentration of guanine is equalled by molar concentration of cytosine.
[A] = [T], i.e., [A] / [T] = 1; [G] = [C], i.e., [G] / [C] = 1
(iii) Sugar deoxyribose and phosphate occur in equimolar proportions.
(iv) A-T base pairs are rarely equal to С—G base pairs.
(v) The ratio of [A+T] / [G+C] is variable but constant for a species (Table 6.2). It can be used to identify the source of DNA. The ratio is low in primitive organisms and higher in advanced ones.
Table 6.2. Base Composition of DNA from Various Sources:
| Species | A | G | С | T | A+T/C+G |
| 1. Man | 30.4 | 19.0 | 19.9 | 30.1 | 1.55 |
| 2. Calf | 29.0 | 21.2 | 21.2 | 28.5 | 1.35 |
| 3. Wheat germ | 28.1 | 21.8 | 22.7 | 27.4 | 1.25 |
| 4. Pea | 30.8 | 19.2 | 18.5 | 30.5 | 1.62 |
| 5. Euglena | 22.6 | 27.7 | 25.8 | 24.4 | 0.88 |
| 6 Escherichia coli | 24.7 | 26.0 | 25.7 | 23.6 | 0.93 |
Structure of DNA:
DNA or deoxyribonucleic acid is a helically twisted double chain polydeoxyribonucleotide macromolecule which constitutes the genetic material of all organisms with the exception of rhinoviruses. In prokaryotes it occurs in nucleoid and plasmids. This DNA is usually circular. In eukaryotes, most of the DNA is found in chromatin of nucleus.
It is linear. Smaller quantities of circular, double stranded DNA are found in mitochondria and plastids (organelle DNA). Small sized DNAs occur in viruses, ф x 174 bacteriophage has 5386 nucleotides. Bacteriophage lambda (Phage X) possesses 48502 base pairs (bp) while number of base pairs in Escherichia coli is 4.6 x 106. A single genome (haploid set of 23 chromosomes) has about 3.3 x 109 bp in human beings. Single-stranded DNA occurs as a genetic material in some viruses (e.g., phage ф x 174, coliphage fd, M13). DNA is the largest macromolecule with a diameter of 2 nm (20Å or 2 x 10-9m) and often having 3 length in millimetres.
It 15 negatively charged due to phosphate groups. It is a long chain polymer of generally several hundred thousands of deoxyribonucleotides. A DNA molecule has two un-branched complementary strands. They are spirally coiled. The two spiral strands of DNA are collectively called DNA duplex (Fig. 6.5).
The two strands are not coiled upon each other but the whole double strand (DNA duplex) is coiled upon itself around a common axis like a rope stair case with solid steps twisted into a spiral. Due to spiral twisting, the DNA duplex comes to have two types of alternate grooves, major (22 Å) and minor (12 Å).
In B-DNA, one turn of the spiral has about 10 nucleotides on each strand of DNA. It occupies a distance of about 3.4 nm (34 Å or 3.4×10-9 m) so that adjacent nucleotides or their bases are separated by a space of about 0.34 nm (0.34×10-9 m or 3.4 Å).
A deoxyribonucleotide of DNA is formed by cross-linking of three chemicals ortho- phosphoric acid (H3PO4), deoxyribose sugar (C5H10O4) and a nitrogen base. Four types of nitrogen bases occur in DNA. They belong to two groups, purines (9-membered double rings with nitrogen at 1,3,7 and 9 positions) and pyrimidines (six membered rings with nitrogen at 1 and 3 positions). DNA has two types of purines (adenine or A and guanine or G) and two types of pyrimidines (cytosine or С and thymine or T).
Depending upon the type of nitrogen base, DNA has four kinds of deoxyribonucleotides —deoxy adenosine 5- monophosphate (d AMP), deoxy guaninosine 5-monophosphate (d GMP), deoxy thymidine 5-monophosphate (d TMP) and deoxy cytidine 5- monophosphate (d CMP).
The back bone of a DNA chain or strand is built up of alternate deoxyribose sugar and phosphoric acid groups. The phosphate group is connected to carbon 5′ of the sugar residue of its own nucleotide and carbon У of the sugar residue of the next nucleotide by (3 ‘—5’) phosphodiester bonds. -H of phosphate and -OH of sugar are eliminated as H20 during each ester formation.
Phosphate group provides acidity to the nucleic acids because at least one of its side group is free to dissociate. Nitrogen bases lie at right angles to the longitudinal axis of DNA chains. They are attached to carbon atom 1 of the sugars by N-glycosidic bonds. Pyrimidine (C or T) is attached to deoxyribose by its N-atom at 1 position while a purine (A or G) does so by N-atom at 9 position.
The two DNA chains are antiparallel that is, they run parallel but in opposite directions. In one chain the direction is 5’→ У while in the opposite one it is 3′ →5′ (Fig. 6.5). The two chains are held together by hydrogen bonds between their bases. Adenine (A), a purine of one chain lies exactly opposite thymine (T), a pyramidine of the other chain. Similarly, cytosine (C, a pyrimidine) lies opposite guanine (G a purine). This allows a sort of lock and key arrangement between large sized purine and small sized pyrimidine.
It is strengthened by the appearance of hydrogen bonds between the two. Three hydrogen bonds occur between cytosine and guanine (C = G) at positions 1’-1’, 2′- 6′ and 6′-2′. There are two such hydrogen bonds between adenine and thymine (A=T) which are formed at positions 1’-3′ and 6′-4′. Hydrogen bonds occur between hydrogen of one base and oxygen or nitrogen of the other base. Since specific and different nitrogen bases occur on the two DNA chains, the latter are complementary.
Thus the sequence of say AAGCTCAG of one chain would have a complementary sequence of TTCGAGTC on the other chain. In other words, the two DNA chains are not identical but complementary to each other. It is because of specific base pairing with a purine lying opposite a pyrimidine. This makes the two chains 2 nm thick.
A purine- purine base pair will make it thicker while a pyrimidine- pyrimidine base pair will make it narrower than 2 nm. Further, A and С or G and T do not pair because they fail to form hydrogen bonds between them. 5′ end of each chain bears phosphate radical while the 3′ end possesses a sugar residue (З’-ОН).
Salient Features of В model of DNA of Watson and Crick:
1. DNA is the largest biomolecule in the cell.
2. DNA is negatively charged and dextrorotatory.
3. Molecular configuration of DNA is 3D.
4. DNA has two polynucleotide chains.
5. The two chains of DNA have antiparallel polarity, 5′ —> 3′ in one and 3′ —> 5′ in other.
6. Backbone of each polynucleotide chain is made of alternate sugar-phosphate groups. The nitrogen bases project inwardly.
7. Nitrogen bases of two polynucleotide chains form complementary pairs, A opposite T and С opposite G.
8. A large sized purine always comes opposite a small sized pyrimidine. This generates uniform distance between two strands of helix.
9. Adenine (A) of one polynucleotide chain is held to thymine (T) of opposite chain by two hydrogen bonds. Cytosine (C) of one chain is similarly held to guanine of the other chain by three hydrogen bonds.
10. The double chain is coiled in a helical fashion. The coiling is right handed. This coiling produces minor and major grooves alternately.
11. The pitch of helix is 3.4 nm (34 A) with roughly 10 base pairs in each turn. The average distance between two adjacent base pairs comes to about 0.34 nm (0.34 x 10-9 m or 3.4 A).
12. Planes of adjacent base pairs are stacked over one another. Alongwith hydrogen bonding, the stacking confers stability to the helical structure.
13. DNA is acidic. For its compaction, it requires basic (histone) proteins. The histone proteins are +vely charged and occupy the major grooves of DNA at an angle of 30° to helix axis.
Sense and Antisense Strands:
Both the strands of DNA do not take part in controlling heredity and metabolism. Only one of them does so. The DNA strand which functions as template for RNA synthesis is known as template strand, minus (-) strand or antisense strand.
Its complementary strand is named nontemplate strand, plus (+) strand, sense and coding strand. The latter name is given because by convention DNA genetic code is written according to its sequence.
(5’) GCATTCGGCTAGTAAC (3′)
DNA Nontemplate, Sense (+) or Coding Strand
(3′) CGTAAGCCGATCATTG (5′)
DNA Template, Antisense, or Noncoding or (-) Strand
(5′) GCAUUCGGCUAGUAAC (3′)
RNA Transcript
RNA is transcribed on 3’→5′ (-) strand (template/antistrand) of DNA in 5 → 3 direction.
The term antisense is also used in wider prospective for any sequence or strand of DNA (or RNA) which is complementary to mRNA.
Denaturation and Renaturation:
The H-bonds between nitrogen bases of two strands of DNA can break due to high temperature (82-90°C) or low or high pH, so that the two strands separate from each other. It is called denaturation or melting. Since A-T base pair has only 2H bonds, the area rich in A-T base pairs can undergo easy denaturation (melting). These areas are called low melting areas because they denature at comparatively low temperature. The area rich in G- C base pairs (called high melting area) is comparatively more stable and dense because three hydrogen bonds connect the G-C bases.
These areas have high temperature of melting (Tm). On melting the viscosity of DNA decreases. The denatured DNA has the tendency to reassociate, i.e., the DNA strands separated by melting at 82-90°C can reassociate and form duplex on cooling to temperature at 65°C. It is called renaturation or annealing.
Denatured or separated DNA strands absorb more light energy than the intact DNA double strand. The increased absorption of light energy by separated or denatured DNA strands is called hyperchromatic effect. The effect is used in knowing whether DNA is single or double stranded.
Palindromic DNA:
DNA duplex possesses areas where sequence of nucleotides is the same but opposite in the two strands. These sequences are recognised by restriction endonucleases and are used in genetic engineering. Given hereunder sequence of bases in one strand (3′ → 5′) is GAATTC. It is same in other strand when read in 5′ → 3′ direction. It is similar to palindrome words having same words in both forward and backward direction, e.g., NITIN, MALAYALAM.
Repetitive DNA:
It is the DNA having multiple copies of identical sequences of nitrogen bases. The number of copies of the same base sequence varies from a few to millions. DNA having single copy of base sequences is called unique DNA. It is made of functional genes. rRNA genes are, however, repeated several times. Repetitive DNA may occur in tandem or interspersed with unique sequences.
It is of two types, highly repetitive and moderately repetitive. Highly repetitive DNA consists of short sequences of less than 10 base pairs which are repeated millions of times. They occur in preeentromeric regions, heterochromatic regions of Y-chromosomes and satellite regions. Moderately repetitive dna consists of a few hundred base pairs repeated at least 1000 times. It occurs in telomeres, centromeres and transposons.
Tandemly repeated sequences are especially liable to undergo misalignments during chromosome pairing, and thus the size of tandem clusters tends to be highly polymorphic, with wide variations between individuals. Smaller clusters of such sequences can be used to characterize individual genomes in the technique of “DNA-finger-printing”.
Satellite DNA:
It is that part of repetitive DNA which has long repetitive nucleotide sequences in tandem that forms a separate fraction on density ultracentrifugation. Depending upon the number of base pairs involved in repeat regions, satellite DNA is of two types, microsatellite sequences (1-6 bp repeat units flanked by conserved sequences) and minisatellite sequences (11-60 bp flanked by conserved restriction sites). The latter are hyper variable and are specific for each individual. They are being used for DNA matching or finger printing as first found out by Jeffreys et al (1985).
Genetic Information:
The arrangement of nitrogen bases of DNA (and its product mRNA) determines the sequence of amino acid groups in polypeptides or proteins formed over ribosomes. One amino acid is specified by the sequence of three adjacent nitrogen bases. The latter is called codon. The segment of DNA which determines the synthesis of complete polypeptide is known as cistron.
In procaryotes, a cistron has a continuous coding sequence from beginning to end. In eucaryotes a cistron contains noncoding regions which do not produce part of gene product. They are called introns. Introns are often variable. The coding parts are known as exons. Cistrons having introns are called split genes.
Coding and Noncoding DNA:
Depending on the ability to form functional or nonfunctional products, DNA has two types of segments, coding and noncoding. In eukaryotes a greater part of DNA is noncoding since it does not form any functional product. They often possess repeated sequences or repetitive DNA. Most of them have fixed positions.
Some can move from one place to another. The mobile sequences are called jumping genes or transposons. In prokaryotes the amount of noncoding or nonfunctional DNA is small. Coding DNA consists of coding DNA sequences. These are of 2 types — protein coding sequences coding for all proteins except histone and nonprotein coding sequences for tRNA, rRNA and histones.
Functions of DNA:
1. Genetic Information (Genetic Blue Print):
DNA is the genetic material which carries all the hereditary information. The genetic information is coded in the arrangement of its nitrogen bases.
2. Replication:
DNA has unique property of replication or production of carbon copies (Autocatalytic function). This is essential for transfer of genetic information from one cell to its daughters and from one generation to the next.
3. Chromosomes:
DNA occurs inside chromosomes. This is essential for equitable distribution of DNA during cell division.
4. Recombination’s:
During meiosis, crossing over gives rise to new combination of genes called recombinations.
5. Mutations:
Changes in sequence of nitrogen bases due to addition, deletion or wrong replication give rise to mutations. Mutations are the fountain head of all variations and evolution.
6. Transcription:
DNA gives rise to RNAs through the process of transcription. It is heterocatalytic activity of DNA.
7. Cellular Metabolism:
It controls the metabolic reactions of the cells through the help of specific RNAs, synthesis of specific proteins, enzymes and hormones.
8. Differentiation:
Due to differential functioning of some specific regions of DNA or genes, different parts of the organisms get differentiated in shape, size and functions.
9. Development:
DNA controls development of an organism through working of an internal genetic clock with or without the help of extrinsic information.
10. DNA Finger Printing:
Hypervariable microsatellite DNA sequences of each individual are distinct. They are used in identification of individuals and deciphering their relationships. The mechanism is called DNA finger printing.
11. Gene Therapy:
Defective heredity can be rectified by incorporating correct genes in place of defective ones.
12. Antisense Therapy:
Excess availability of anti-mRNA or antisense RNAs will not allow the pathogenic genes to express themselves. By this technique failure of angioplasty has been checked. A modification of this technique is RNA interference (RNAi).