This article provides useful notes on the Biosynthesis of DNA Replication!
The biosynthesis of deoxyribonucleic acid (DNA) in vitro through polymerization of nucleoside triphosphates in the presence of the appropriate enzyme was first successfully completed by Kornberg, and for this he received the Nobel Prize in 1959.
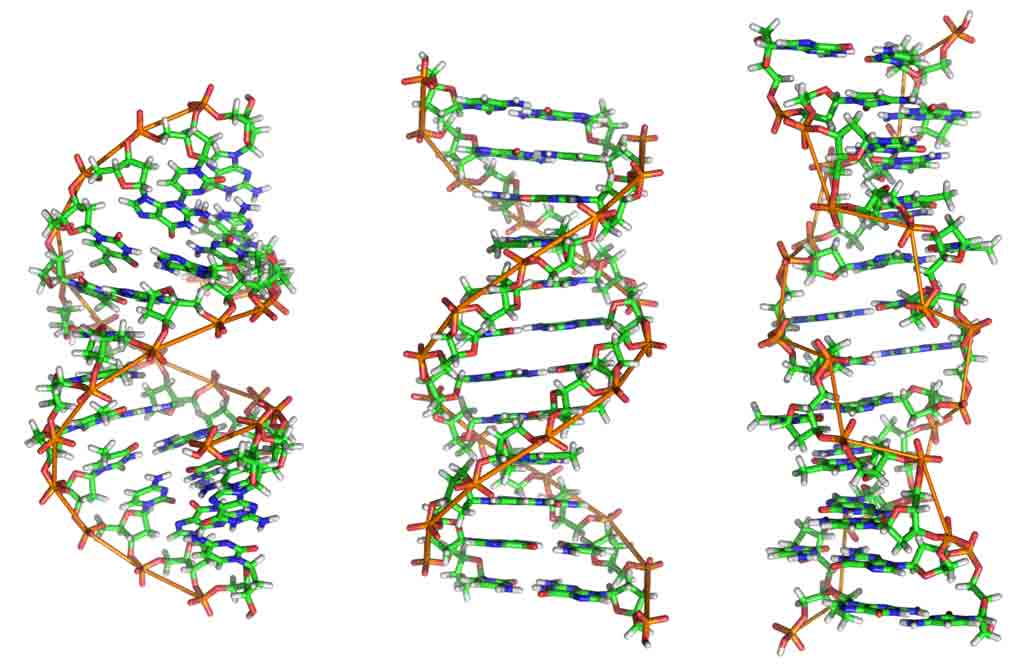
The nucleic acid DNA consists of four kinds of nucleotides: units of purine or pyrimidine attached to a phosphoric acid residue through a pentose sugar (Freifelder, 1978).
In DNA the nucleotides attach to one another in strands, the phosphoric acid and pentose forming the backbone of the spiralling structure of Watson-Crick DNA model. The two complementary DNA strands are held together by relatively weak hydrogen bonds between their respective complementary purine and pyrimidine bases-thymine (T) to adenine (A), and guanine (G) to cytosine (C).
Each polynucleotide chain serves as a template for the synthesis of a new DNA molecule. DNA replication follows the base pairing rules by which A pairs only with T, and G with C, and as a consequence of this, each daughter molecule is an exact replica of the parent molecule.
Contents
- [I] DNA replication is semiconservative:
- [II] DNA synthesis is discontinuous on one strand:
- [III] Replication requires Helix-destabilizing proteins and DNA topoisomerases:
- [IV] Copying of RNA into DNA:
- [V] Bidirectional replication of bacterial DNA:
- [VI] Multiple origins of replication of eukaryotic chromosomes:
[I] DNA replication is semiconservative:
Watson-Crick proposed that one of the strands of each daughter DNA molecule is newly synthesized, whereas the other is derived from the parent DNA molecule. This distribution of parental atoms is called semiconservative.
A critical test of this hypothesis was carried out by Matthew, Meselson and Franklin Stahl. The parent DNA was labelled with 15N, the heavy isotope of nitrogen to make it denser than ordinary DNA. This was accomplished by growing E. coli for many generations in a medium that contained 15NH4Cl as the sole nitrogen source.
The bacteria were abruptly transferred to a medium that contained 14N, the ordinary isotope of nitrogen. (DNA containing 15N is more dense than the DNA containing 14N.) The DNA was isolated and its density was determined by ultracentrifugation on a cesium chloride gradient.
It was found that after the first division cycle there is only one DNA peak corresponding to the heavy-light (HL) hybrid molecule (i.e., one strand is labelled with 14N and the other with 15N). At the second generation two peaks of DNA appear-one in which the two DNA strands contain 14N (LL) and the other still corresponding to hybrid molecules.
[II] DNA synthesis is discontinuous on one strand:
DNA is synthesized by enzymes called DNA polymerases, which are required not only for DNA replication but also for DNA repair. DNA polymerases of both prokaryotic and eukaryotic origin require for their function the presence of a DNA template, a primer (RNA or DNA, but only RNA is used in vivo), and the four deoxynucleoside triphosphates (dATP, dTTP, dGTP and dCTP), which are building blocks of DNA.
DNA polymerases can only add nucleotides to the 3′ OH group of a preexistent primer, and DNA synthesis can only proceed in the 5’→ 3′ direction. Because both strands of a DNA molecule are antiparallel, i.e., run in opposite directions, this creates a problem.
When the two strands unwind, one of them will be facing the DNA polymerase in the correct direction (5′ →3′), but the other strand will have an unfavourable orientation (3′ →5′). Thus the DNA synthesis in one segment proceeds in a discontinuous way, synthesizing short segments of DNA (always in 5′ → 3′ direction). These segments are then joined together by the action of a DNA ligase.
Okasaki fragments:
The short discontinuous DNA segments were discovered by T. Okasaki who exposed bacteria to 3H-thymidine for a few seconds and found fragments of DNA 1000 to 2000 nucleotides. These pieces are called Okasaki fragments after their discoverer. In eukaryotes these are 200 nucleotides long.
DNA polymerases can only elongate a primer molecule; they cannot start a new chain by themselves. A short segment of RNA is synthesized and it acts as the primer. This RNA segment is later removed by repair enzymes and the gap is filled by DNA polymerase, and then joined to the neighbouring Okasaki fragments by a DNA ligase.
[III] Replication requires Helix-destabilizing proteins and DNA topoisomerases:
Replication machinery of DNA consists of many proteins which act on the replication fork.
1. Helix-destabilizing proteins:
These proteins bind to single-stranded DNA and thus help to open up the double helix at the replication fork. Both strands of DNA helix are held together by weak hydrogen bonds, and these bonds break up by helix-destabilizing proteins.
2. DNA topoisomerases:
These enzymes can change the shape of DNA. Some of these enzymes can introduce supercoils, unwind DNA molecules.
The simplest DNA topoisomerases (type I) introduce a cut on only one of the DNA Strands. This allows the molecule to rotate around phosphodiester bond on the opposite strand. This is important during DNA replication, because as the two strands of DNA are separated at the replication fork, the rest of DNA tends to ‘wind up’ and accumulate tension.
Type 1 topoisomerases do not require ATP to work. They bind covalently to the cut DNA strand storing the chemical energy of the Phosphodiester Bridge, which they then use to reseal the strand after the tension has been removed from the DNA.
Type II DNA toposiomerases can cut or untangled (and subsequently reseal) both strands of a DNA molecule. It requires ATP.
[IV] Copying of RNA into DNA:
Mostly there is unidirectional flow of information, i.e.,
replication DNA → transcription RNA → translation Protein
But sometimes RNA can be copied into DNA. In other words RNA may be transcribed into DNA and this is done by reverse transcriptase, an RNA-dependent DNA polymerase that is able to synthesize DNA on an RNA template.
Replication DNA, ↔ RNA → Protein
Tumor-producing viruses containing an RNA genome, called retroviruses have been found to act as templates for the synthesis of DNA. Tumor virion contains the reverse transcriptase by which an RNA/DNA hybrid molecule can be produced.
In this way viral genes can be integrated into the genome of the host cell. Most higher eukaryotes contain retroviruses within their genome and reverse transcriptase, which can copy normal cellular RNAs into DNA that can be subsequently integrated in the genome.
[V] Bidirectional replication of bacterial DNA:
E. coli (prokaryote) has a circular chromosome (DNA), which upon unfolding has a length of about 1.1 mm. E. coli chromosome is made of a single molecule of DNA and is attached at one point to the cell membrane.
In E. coli (as in eukaryotes) DNA replication proceeds bidirectionally starting from a fixed origin of replication. The entire replication of E. coli chromosome takes 30 minutes. From the initiation point of the circular chromosome, two replication forks proceed bidirectionally. At a certain stage, also starting from the same point of origin, two new replication forks are initiated, thus producing four forks which advance bidirectionally.
[VI] Multiple origins of replication of eukaryotic chromosomes:
Eukaryotic chromosomes contain a large amount of DNA, i.e., Amphiuma contain 2.5 meters of DNA in a single molecule; human chromosomes have 7 cm. If these huge molecules were replicated from a single origin of replication, as in E. coli, the S phase of cell division would be too much long. Eukaryotic cells solve this problem by having multiple replication initiation sites in chromosome.
(1) Two adjacent replicons prior to replication;
(2) Replication started towards right;
(3) Replication started towards left also;
(4) Replication completed in both.
In 1968 Huberman and Riggs demonstrated that eukaryotic 5MA к as tandem units of replication, called replicons. The average replicons are 30 µm long, and there are about 30,000 of them per haploid mammalian genome. Each chromosome may have several thousand origins of replication.
Each replication fork progresses at a speed of 50 nucleotides per second, which is 10 times slower than in E. coli. Replication forks proceed in opposite directions from each origin (O) until they reach the terminal point at which they meet the neighbouring replicon. The points of termination are not fixed, and they correspond to the point at which two growing forks converge.