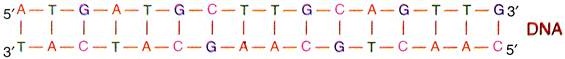Let us make an in-depth study of the deoxyribonucleic acid. After reading this article you will learn about: 1. Deoxyribonucleic Acid (DNA) 2. Structure of DNA 3. Functions of DNA 4. Packaging of DNA and 5. Physical Properties of DNA.
Contents
Deoxyribonucleic Acid (DNA):
Deoxyribonucleic acid, also abbreviated as DNA, is the principal informational macromolecule of the cell, which stores, translates and transfers the genetic information. In the prokaryotes, the DNA is found mostly in the nuclear zone. In eukaryotes it is found in the nucleus, mitochondria and chloroplast. The present understanding of the storage and utilization of the cell’s genetic information is based upon the discovery of the structure of DNA by Watson and Crick in 1953.
Structure of DNA:
Double Helical Structure of DNA (Watson and Crick Model):
The three dimensional structure of DNA as proposed by Watson and Crick and the recent advances in it are summarized here:
1. DNA is made of two helical chains coiled around the same axis, to form a right-handed double helix.
2. The two chains in the helix are anti-parallel to each other, i.e., the 5′-end of one polynucleotide chain and the 3′-end of the other polynucleotide chain is on the same side and close together.
Double helical structure of DNA
3. The distance between each turn is 3.6 nm (formerly 3.4 nm).
4. There are 10.5 nucleotides per turn (formerly 10 nucleotides).
5. The spatial relationship between the two strands creates major and minor grooves between the two strands. In these grooves some proteins interact.
6. The hydrophilic backbones of alternating deoxyribose and negatively charged phosphate groups are on the outside of the double helix.
7. The hydrophobic pyrimidine and purine bases are inside the double helix, which stabilizes the double helix of the DNA.
8. The double helix is also stabilized by inter-chain hydrogen bond formed between a purine and pyrimidine base.
9. A particular purine base, pairs by hydrogen bonds, only with a particular pyrimidine base, i.e., Adenine (A) pairs with Thymine (T) and Guanine (G) pairs with Cytosine (C) only.
10. Two hydrogen bonds pairs Adenine and Thymine (A = T), whereas three hydrogen bonds pairs Guanine and Cytosine (G ≡ C).
11. The base pairs A = T and G ≡ C are known as complementary base pairs.
12. Due to the presence of complementary base pairing, the two chains of the DNA double helix are complementary to each other.
Hence the number of A’ bases are equal to the number of T’ bases (or ‘G’ is equal to ‘C) in a given double stranded DNA.
13. One of the strands in the double helix is known as sense strand, i.e., which codes for RNA/proteins and the other strand is known as antisense strand.
Different Structural Forms of DNA:
The DNA molecules exist in four different structural forms or organizations under different physiological conditions or in different cells or at different points in the same DNA.
Functions of DNA:
The base sequence of the DNA constitutes the informational signal called the genetic material. This nucleotide base sequence enables the DNA to function, store, express and transfer the genetic information. Hence it programs and controls all the activities of an organism directly or indirectly throughout its life cycle.
(a) DNA stores the complete genetic information required to specify (form) the structure of all the proteins and RNA’s of each organism.
(b) DNA is the source of information for the synthesis of all cellular body proteins. Some of the proteins are structural proteins and some are enzymes. These enzymes arrange micro-molecules to form macromolecules. These macromolecules are arranged to form supra-molecular complexes or cell organelles which associate to form cells. These cells group to form tissues which in turn form different organs of a body, specifically peculiar to that organism during foetal development, growth and repair. Hence DNA programs in time and space the orderly biosynthesis of cells and tissue components.
(c) It determines the activities of an organism throughout its life cycle, i.e., the period of gestation, birth, maturity, senescence and death.
(d) It determines the individuality and identity of a given organism.
(e) It duplicates (replicates to form two daughter DNA) itself and transfers one of the copy to the daughter cell during cell division, thus maintaining the genetic material from generation to generation.
Packaging of DNA:
Packaging of DNA within the Cells:
The length of DNA molecules existing in a particular cell is much longer than the long dimensions of the cell or the organelle where they exist. The contour length (i.e., its helix length) of a double stranded DNA can be calculated from the molecular weight, presuming the average molecular weight of each nucleotide pair to be about 650 Da and there is one nucleotide pair for every 0.36 nm of the duplex.
Accordingly the length of the smallest DNA is 1938.96 nm, belonging to the ɸX174 virus (in duplex form), whose particle long dimension is 25 nm. On the other hand the total contour length of the entire DNA in a single human cell is about 2 metres and the cell nucleus is just 5-10 nm in diameter. These long DNA molecules are very tightly compacted, so as to fit into the cell.
This packing is possible due to DNA getting further coiled into different fashions. The linear double helical DNA called the relaxed DNA, bends or twists upon its own axis, which is called as DNA coiling. This coiled DNA further coils upon itself to form the DNA supercoil, just like the telephone cord wire from the base of the phone to the receiver.
The degree of DNA supercoiling depends upon the type of the cell/organelle to be packed and is coiled in such a manner that DNA can easily be accessible to the enzymes/proteins for all of its functions like replication and transcription.
There are possibilities of two types of DNA supercoiling:
(a) Solenoidal, wherein the DNA coils in a spherical fashion upon itself and
(b) Plectonemic, wherein the DNA coils back upon its reverse length in the form of pleats. Further the supercoiling and packing of DNA differ in the prokaryotes (i.e., those lacking a true nuclear envelope) and in the eukaryotes (i.e., those having a nuclear membrane).
(a) Packaging of viral DNA:
Though the viral DNA is much smaller than a bacterial or eukaryotic DNA, its contour length is much bigger than the long dimensions of the viral particle in which they are found. The DNA of bacteriophage T2 is 3500 times its particle diameter. The long dimensions of different viral particle/the contour length of their DNAs respectively in nanometers is T2-210/65520; Lamdaphage-190/17460; Ti-78/14376 and φX174 (in duplex form)-25/1938. In order to get themselves packed within the particle, most of these viral DNAs are linked covalently by the ends and, therefore, form an endless belt and thus become circular. Some of the single stranded DNA (φX174) becomes double stranded and circular.
(b) Packaging of bacterial DNA:
The length of E.coli cell is 2 micro-meters and its complete DNA molecule (the complete DNA molecule of an organism is called the chromosome) is 1.7 mm long, which exists as a single, covalently closed double stranded circular molecule, coiled and packed within nucleon of the cell.
This circular chromosome is organized in a scaffold-like structure, which folds the chromosome into looped domains. These domains are further coiled around some basic proteins, called Hu proteins (Mw = 10 000). In addition to the nucleosomal DNA the bacteria contains some small circular supercoiled non-chromosomal DNA-called plasmids.
(c) Organization and packing of eukaryotic DNA:
All the chromosomal DNA of an eukaryotic cell is embedded in a membranous cellular organelle called the nucleus. The eukaryotic DNA, in the nucleus is linear and not circular. In the non-dividing resting cell all the DNA of the cell forms a fine filamentous network in the nucleus called the chromatin. During cell division the chromatin network is subdivided into defined number and shaped chromosomes, their diploid number (pairs) depends upon the species of organism.
The normal chromosome number in humans is 46 (23 pairs). Each chromosome has a central axis called the centromere, from which two arms of DNA project out and each is referred to as chromatid. Each chromosome differs in size and shape within a given organism.
The 2 metre long eukaryotic human cell DNA is to be packed in the cell of about 5-10 micrometer in diameter. In order to facilitate its package, the helical DNA molecule is bound, tightly around beads of basic proteins called histones, which are spaced at regular intervals. The complexes of histones and DNA are called nucleosomes. Each nucleosome contains eight histone molecules, two copies each of H2A, H2B, H3 and H4 winding 146 base pairs of DNA.
Between the two nucleosomes there is a spacer DNA of 54 base pairs with a single histone molecule (H1). Wrapping DNA around a nucleosome compacts it to about seven-fold. These nucleosomes are organized very close together to form a structure, simply called 30 nm fibre. This provides 100-fold compaction of DNA. The 30 nm fibre then forms plectonemic pleats, called loops.
Six such loops are bounded by scaffold attachment to proteins (histone- H1/topoisomerase-II) to give rise to a cluster of loops called rosette. 30 such rosettes bunch together to form a single coil, 10 such coils (like phone cord) forms a chromatid and the two chromatids are linked together by a highly repetitive base sequence rich in AT base pairs called satellite DNA, which is the centromere. Two chromatids with a centromere form a chromosome.
Physical Properties of DNA:
(a) Denaturation:
When DNA is subjected to extremes of pH or temperatures above 80 to 90 degree centigrade, it gets denatured and the double helical structure is unfolded due to disruption of hydrogen bonds between the bases and the hydrophobic interactions of the bases. Finally, the two strands separate completely from each other. This is melting of DNA. The temperature at which a given DNA is denatured to about 50% is known as TM.
Different DNA melts at different temperatures, which depends upon the G ≡ C content of that DNA. Higher the G ≡ C content, higher is the melting temperature (TM) and vice-versa. When the temperature or pH is slowly brought back to normal biological range, the two strands will automatically rewind or anneal and will again form the same double helical structure. If the temperature is suddenly cooled down, then the two strands remain separated and exist as single strands.
(b) Buoyant Density:
When DNA is centrifuged at high speeds in a concentrated solution of caesium chloride-(CsCI), the CsCl will form a density gradient (ascending) and the DNA will remain stationary or buoyant at a point in the tube where its density is equal to the density of CsCI at that point. Different DNA will have different densities, which again depend upon the GsC content of that DNA. Higher the G = C content, higher is the buoyant density of that DNA and vice versa.
Measurement of these two characters, viz., melting temperature and buoyant density will enable us to calculate the proportions of G ≡ C and A = T pairs in that DNA, which indirectly helps in deducting the gene sequence.