The following points highlight the eight important cytoplasmic inclusions found in bacteria. The cytoplasmic inclusions are: 1. Ribosomes 2. Polyphosphates 3. Poly-β-hydroxybutyrate 4. Glycogen 5. Gas Vacuoles 6. Magnetosomes 7. Sulfur Globules 8. Carboxysomes.
Cytoplasmic Inclusion # 1. Ribosomes:
Ribosomes (Fig. 5.20) in bacteria (prokaryotes) are small granular bodies of 10-20 nm in diameter freely lying in the cytoplasm and composed of ribosomal ribonucleic acid (rRNA) and proteins. Bacterial ribosomes are thought to contain about 80-85% of the bacterial RNA.
Sometimes, they are found in small groups called polyribosomes ox polysomes, which are formed when several ribosomes begin to translate a single mRNA molecule. Generally, the ribosomes are a few hundred in number in each bacterial cell, but when the cell undertakes active protein synthesis, they increase in number to as many as 15,000-20,000 per cell about 15% of the cell mass.
1. Subunits:
Each ribosome has sedimentation coefficient of 70S and a mass of 2.8 x 106 daltons and is made up of two subunits of 50S and 30S, each subunit consisting of roughly equal amounts of rRNA and protein. The number 30S, 50S, and 70S refer to Svedberg units, which are units of sedimentation coefficient of ribosome subunits (30S and 50S) or intact ribosomes (70S) when subjected to centrifugal force in an ultracentrifuge.
Ribosomes are functional only when the two subunits are combined together. The association and dissociation of two subunits of ribosomes depend on the concentration of Mg++ ions.
Each 50S subunit (mass of 1.8 x 106 daltons) contains one molecule of 23S rRNA (having approximately 3200 nucleotides), one molecule of 55 rRNA (having only about 120 nucleotides) and 34 different proteins designated as L1 to L34; while the 30S subunit (mass of 0.9 x 106 daltons) contains one molecule of 16 rRNA (having approximately 1540 nucleotides) and 21 different proteins designated as S1 to S21.
2. Models:
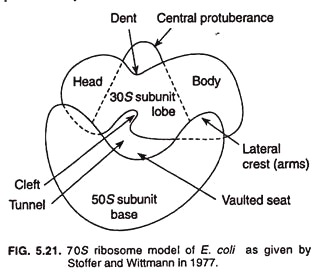
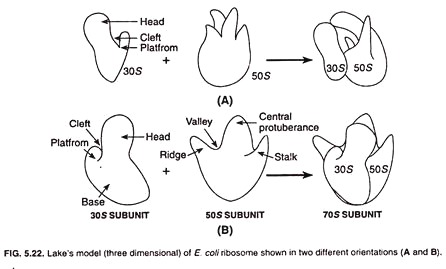
Two types of models have been presented to explain bacterial ribosome structure. These are Stoffer and Wittmann’s model (1977) and Lake’s model (1981).
Stoffer and Wittmann’s Model:
Stoffer and Wittmann (1977) presented their model of bacterial ribosome which explains quasi symmetrical structure of ribosome (Fig. 5.21). Structurally 30S sub unit is elongated, slightly bent, and prolate shape, and is divided into two parts, namely, head (smaller) and body (larger) by a hollow or cleft. The 50S subunit may be rounded, reniform (kidney-shaped), circular, or maple leaf structure.
Lake’s Model:
James A. Lake presented a new model for ribosome structure in 1981. According to this model the 30S subunit of ribosome appears asymmetrical and consists of a head, a base, and a platform. The head and platform are separated from the base with the help of a cleft.
The 50S subunit is also asymmetrical and comprises of a ridge, a central protuberance, and a stalk. The ridge and central protuberance are separated by a valley (Fig. 5.22). The ridge and talk are laterally projected and incline at an angle of about 50° from the central protuberance.
3. Functions:
As in eukaryotes, ribosomes are the sites of protein synthesis in bacteria. Several antibiotics such as streptomycin, neomycin, tetracyclines, and chloramphenicol specifically inhibit protein synthesis by attacking ribosomes. Protein synthesis involves a complex cycle in which the various ribosomal components play specific roles.
The ribosome plays a key role in the translation process, bringing together mRNA and aminoacyl tRNAs. There are three sites on the ribosome: the acceptor site, where the charged tRNA first combines; the peptide site, where the growing polypeptide chain is held; and exite site.
During each step of amino acid addition, the ribosome advances three nucleotides (one codon) along the mRNA and the tRNA moves from the acceptor to the peptide site. Termination of protein synthesis takes place when a nonsense codon, which does not encode an amino acid, is reached.
Cytoplasmic Inclusion # 2. Polyphosphates (Volutin Granules or Metachromatin Granules):
Many bacteria and microalgae accumulate inorganic phosphates in the form of granules of polyphosphates. Polyphosphate is a liner polymer of orthrophosphates joined by ester bonds (Fig. 5.23).
Because they were first described in Spirillum volutans and because they bring a about metachromatic effect (i.e., appear red or a different shade of blue when stained with methylene blue or toluidine blue dyes), they have also been given the name ‘volutin granules’ and ‘metachromatin granules’, respectively.
These granules are composed of polymetaphosphate and are common in diphtheria bacillus and in certain lactic acid bacteria. The chemical structure of polyphosphate is shown in fig. 5.24.
These granules refractive and hence arc easily observable under light microscope. The polyphosphates represent intracellular phosphate reserve when nucleic acid synthesis does not occur, and when the letter starts, the polyphosphate granules are degraded and used as sources of phosphate for nucleic acids.
Polyphosphates are also used as source of phosphate for phospholipids. In some cells the polyphosphates act as an energy reserve and can serve as energy source in reactions.
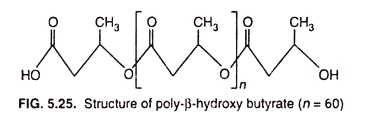
Cytoplasmic Inclusion # 3. Poly-β-hydroxybutyrate (PHB):
Poly- β -hydroxybutyrate (PHB), one of the most common inclusion bodies in bacteria, is a lipid formed from β -hydroxybutyrate monomers (units) joined by easter-linkages between the carboxyl and hydroxyl groups of adjacent molecules resulting in long PHB polymer (Fig. 5.25) which aggregate into granules of around 0.2-0.7 µm in diameter.
The length of the monomer in the polymer can vary considerably, from a short as C4 to as long as C18 in certain bacteria.
Poly-β hydroxybutyrate granules are readily stained with Sudan black for light microscopy and are clearly visible in the electron microscope (Fig. 5.26).
PHB is accumulated by aerobic and facultative bacteria when the cells are deprived of oxygen and must carry out fermentative metabolism. On return of aerobic conditions, PHB, which is a long-term energy storage, is used as an energy and carbon source and incorporated into the oxidative metabolism.
Some bacteria produce co-polymers of PHB often referred to as poly-β-hydroxy-alkanoate (PHA). The latter can be thermo-plastically moulded and used as new plastics that shows advantage over conventional plastics (polypropylene or polyethylene) of being biodegradable.
However, a copolymer containing approximately equal amounts of poly-β-hydroxybutyrate (PHB) and poly-β- hydroxyvalerate (PHV) has had the greatest market success thus far. But, since they are more cost-effective, the conventional petroleum-based plastics still make up virtually the entire plastics market today.
Cytoplasmic Inclusion # 4. Glycogen:
Glycogen (Fig. 5.27) like PHB, is another storage product formed by prokaryotes. It is a polymer of glucose units composed of long chains formed by α(1→ 4) glycosidic bonds and branching chains connected to them by a(1 → 6) glycosidic bonds.
Glycogen is dispersed more evenly throughout the cytoplasmic matrix as small (about 20 – 100 nm in diameter) and is a storage reservoir tor carbon and energy. Glycogen is also known as ‘animal starch’ and, besides prokaryotes, is found in fungi.
Cytoplasmic Inclusion # 5. Gas Vacuoles:
Gas vacuoles, the most remarkable organic inclusion bodies, are formed as a result of the aggregation of enormous number of small, hollow, cylindrical structures called gas vesicles. These structures confer buoyancy on cells by decreasing their density and live a floating existence within the water column of lakes and the oceans.
Each gas vacuole appears about 75 nm in diameter with conical ends and about 200-1,000 nm in length. The most dramatic instances of floatation due to gas vacuoles are seen in cyanobacteria that form massive accumulations (blooms) in lakes.
Gas vacuoles also characteristically occur in many aquatic bacteria such as purple and green photosynthetic ones, and a few non-photosynthetic aquatic bacteria such as Halobacterium and Thiothrix.
Bacteria possessing gas vacuoles can regulate their buoyancy to float at the depth necessary for proper light intensity, oxygen concentration, and nutrient levels. They descend by simply collapsing gas vesicles and further float upward when new gas vesicles are formed and join them.
Each gas vesicle is a spindle-shaped, single membrane-bound gas-filled structure made of protein; the protein subunits assemble to form the wall of the gas vesicle which encloses the hollow cylinder and is impermeable to water but freely permeable to atmospheric gases.
Two different proteins, GvpA and GvpC (Fig. 5.28), compose the gas vesicle wall. GvpA composes 97% of total gas vesicle protein and is the major gas vesicle protein.
It is a small highly hydrophobic and very rigid protein. The rigidity of the gas vesicle wall is essential for the structure to resist the pressures exerted on it from outside. GvpC, the protein in minor amount of 3%, functions to strengthen the wall of the gas vesicle.
Cytoplasmic Inclusion # 6. Magnetosomes:
Magnetosomes are the inorganic inclusion bodies of iron usually in the form of intracellular chains of magnetite (Fe3O4). Some species from sulfidic habitats possess magnetosomes containing greigite (Fe3S4) and pyrite (FeS2). Magnetosome (Fig. 5.29) containing bacteria are called magnetotactic bacteria (e.g., Aquaspirillum magnetotacticum).
Magnetosomes vary in shape from square to rectangular to spike-shaped as their morphology is species-specific. They are around 40 to 100 nm in diameter and bounded by a monolayer membrane made up of phospholipids, proteins, and glycoproteins.
Magnetosome membrane is a non-unit membrane similar to that surrounding granules of poly- β-hydroxybutyrate (PHB) and its proteins probably play a role in precipitating F3+ as Fe3O4 in the developing magnetosome.
Most of the magnetotactic aquatic bacteria grow best at very low O2 concentrations the main function of magnetosomes is probably to guide such bacteria toward the sediment where O2 concentration is lower. Magnetotacic bacteria exhibit magnetotaxis, the process of orienting and migrating along earth’s magnetic field lines, and hence are referred to as the living magnets.
For convenience, magnetotactic bacteria in the Southern hemisphere use their magnetosome chain to determine southward and downward directions and swim down to nutrient-rich sediments or locate the optimum depth in fresh water and marine habitats. Magnetotactic bacteria in Northern hemisphere orient northward and downward for the same purpose.
Despite magnetotactic bacteria, magnetosomes also occur in the heads of birds, dolphins, tuna, green turbles, and other animals, presumably to aid navigation. Magnetotactic bacteria and animals therefore share more in common behaviourally than previously thought.
Cytoplasmic Inclusion # 7. Sulphur Globules:
Sulphur globules (Fig. 5.30) are present in the bacterial cells growing In H2S rich environment such as photosynthetic purple sulfur bacteria and filamentous non-photosynthetic bacteria (Beggiatoa and Thiothrix). These bacteria oxidize H2S into elemental sulfur (H2S → S°) which accumulates inside the cell in visible sulfur globules.
These sulfur globules of elemental sulfur remain until the H2S source is reduced. In the latter condition the stored sulfur in these granules is oxidized to sulfate (S° → SO42-) and the globules slowly disappear.
It is reported that the sulfur globules occur in the periplasm rather than the cytoplasm of the bacterial cell. The periplasm expands outwards to accommodate the globules and contracts when the sulfur of the globules is oxidized.
Cytoplasmic Inclusion # 8. Carboxysomes:
Carboxysomes are polyhedrical bodies surrounded by thin, non-unit membrane and range about 100 nm in diameter. They contain, apart from a little DNA, the enzyme ribulose-1, 5- bisphosphate carboxylase (RUBISCO) in a paracrystalline arrangement.
It is thought that carboxysomes are a mechanism to increase the amount of RUBISCO in the bacterial cell to allow for more rapid CO2 fixation without causing any effect on the osmolarity of the cytoplasm; the osmotic pressure of the cytoplasm is not affected as the carboxysome is insoluble.
Photoautotrophic (cyanobacteria) and chemolithoautotrophic (sulfur bacteria, nitrifying bacteria) that use Calvin cycle for CO2 fixation produce carboxysomes. The latter do not occur in facultative autotrophic bacteria (photoorganoheterotrophic), which grow either as autotrophs or as heterotrophs. Thus, the carboxysomes appear to be an evolutionary adaptation to bacteria under strict autotrophic environment.