In this article we will discuss about:- 1. Recombination in Neurospora 2. Gene Mapping in Neurospora 3. Genotypic Control.
Recombination in Neurospora:
The fungus Neurospora crassa has some advantages for study of crossing over. In the vegetative phase of its life cycle, there is growth of filamentous, coenocytic hyphae to form the haploid mycelium. Sexual reproduction is achieved by coming together of two hyphae of proper mating type and their nuclei fuse to form a diploid zygote (Fig. 8.11).
The zygote starts enlarging into an elongated structure called ascus and the zygote nucleus starts dividing meiotically. The four products of meiosis are arranged in a single linear row inside the ascus. Each one of these undergoes a mitotic division so that 8 nuclei are formed which develop a wall and become ascospores.
What makes Neurospora interesting for a geneticist is the fact that the ascospores are linearly ordered in the same sequence as the chromatids were on the meiotic metaphase plate. It is therefore possible to recover all the four products of a single meiosis and analyse them, as also each chromatid from a tetrad. This is called tetrad analysis.
Neurospora thus presents a direct way of demonstrating recombination which in higher organisms can be inferred on a statistical basis from progeny counts. A still further advantage is the use of centromere as a marker for determining map distances.
When the segregation of a single pair of alleles is analysed, the ascus of Neurospora shows two arrangements of the eight ascospores: 4: 4 ratio resulting from first division segregation; and 2:2:2:2 ratio resulting from second division segregation (Fig. 8.12).
First Division Segregation:
The results of a cross between a normal (al+) and an albino (al) strain of Neurospora are shown in Fig. 8.12. If the 8 ascospores are removed from the ascus and each is grown separately, it is found that 4 ascospores produce mycelium of normal type and 4 of albino type.
This is due to first division segregation explained as follows: during anaphase I of meiosis in the zygote, one homologue carrying al+al+ segregates from the second homologue which has the other allele alal and both move to opposite poles.
Their products are recovered in a 4: 4 ratio after meiosis II (and after mitosis) because no crossing over has occurred between the gene and the centromere. This situation results when the gene is located close to the centromere. Therefore, if a large number of asci are found to exhibit 4: 4 ratio, it indicates that the gene locus in question is close to the centromere.
Second Division Segregation:
As shown in Fig. 8.12 if crossing over occurs between the said gene and the centromere, then one chromatid of each homologue will carry al+ and the other al. Therefore al+ and al will not be able to segregate from each other at anaphase of first meiosis when the two homologues separate to the two poles.
It is only at anaphase of second meiosis when centromeres divide and chromatids separate from each other to the two poles that al+ and al will segregate from each other. This is called second division segregation and produces a ratio of 2al+: 2al: 2al+: 2al, which also indicates that crossing over has occurred between the gene locus and centromere.
Gene Mapping in Neurospora:
In Neurospora the centromere is a marker for determining map distances. For detecting linkage and map distances, the frequency of crossing over is determined from the number of asci showing second division segregation. If there is one crossover, the resulting ascus shows 50% of ascospores with parental combinations and 50% with re-combinations.
Suppose in a cross involving a pair of alleles 30% of asci show second division segregation. This shows that 30% of zygotes had crossing over during meiosis and 70% did not. Since there are four chromatids in each tetrad, the 50% asci have resulted from 30 x 4 = 120 original chromatids in meiosis. When there is crossing over only two of the four chromatids are involved in an exchange.
Therefore only half of the 120 chromatids i.e., 60 are crossover chromatids, the remaining 60 chromatids are non-crossover chromatids. It was also stated above that 70% of zygotes did not have crossing over, which means that 70 x 4 = 280 are non-crossover chromatids. The actual number of non-crossover chromatids is larger because the 30% asci showing second division segregation also have 60 non-crossover chromatids.
The exact number is therefore 280 + 60 = 340. Therefore, of the original 100 tetrads or asci 340 are non-crossover chromatids and 60 are crossover chromatids. Since 100 tetrads or asci also mean 400 chromatids, the percentage of crossover chromatids is (60/400) x 100 = 15%.
From this we can conclude that there was 15% crossing over between the gene and the centromere. We can also say that the gene in question is 15 map units apart from the centromere. Because the centromere itself serves as a marker, in Neurospora it is possible to map a single gene pair. It is also called a two-point cross.
It follows that the method of detecting gene linkage in fungi is basically similar to that for diploids. The main feature in all cases consists in comparing the frequency of parental types to recombinant types. If there is a significant reduction in the frequency of recombinant types from the frequencies expected on the basis of independent assortment, we can consider linkage.
Whereas in Neurospora the meiotic products occur in an ascus in a linear order (ordered tetrads) this is not true for other fungi in Ascomycetes which have unordered tetrads. Let us examine a cross involving two linked genes + +/ab in a fungus having unordered tetrads.
The zygote (++/ab) undergoes meiosis, and depending upon the occurrence of crossover, the tetrads are of following three types:
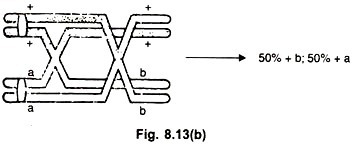
(a) Parental Ditype (PD) (Fig. 8.13a):
If a crossover does not occur between these two loci, or if a two strand double crossover occurs between them, the resulting meiotic products will be of two kinds, both resembling parental combinations, and appear in equal frequency (1 + +; 1 ab). Such a tetrad is called parental ditype (PD).
(b) Non-parental Ditype (NPD) (Fig. 8.13 b):
If a four strand double crossover occurs between the two genes, two kinds of products are formed, both being re-combinations. Such a tetrad is called a non-parental ditype (NPD).
(c) Tetra-type (TT) (Fig. 8.13 c):
This is produced either by a single crossover or a three- strand double crossover (of two types) between the two genes.
Whenever the number of parental di-types and non-parental di-types are significantly unequal, linkage between the two genes must be considered.
For determining the amount of recombination between the two genes, the following formula is used:
Recombination Frequency = NPD + 1/2 TT/Total number of tetrads
Genotypic Control of Neurospora:
In some cases at least recombination itself is under the control of genes. In Neurospora crassa several recombination genes control frequency of recombination either between genes or within genes. Thus a recessive gene rec-1 controls frequency of recombination at the his-1 locus. Both rec-1 and his-1 are on the same chromosome.
In maize chiasma formation can be entirely suppressed by a recessive gene as present on chromosome 1. There are quite a few examples known of single genes as well as polygenes which can cause variation in frequency and distribution of chiasma in one or more chromosomes.