In this article we will discuss about:- 1. Meaning of Human Genome Project 2. History of Human Genome Project 3. Aims 4. Size 5. Mapping 6. Sequencing 7. Outcome.
Contents:
- Meaning of Human Genome Project
- History of Human Genome Project
- Aims of Human Genome Project
- Human Genome Size
- Mapping Human Genome
- Sequencing Human Genome
- Outcome of Human Genome Project
1. Meaning of Human Genome Project:
The Human Genome Project (HGP) is an International collaborative research programme which started in 1990 and completed in 2003, whose goal was the complete mapping and understanding of the three billion DNA subunits (bases), and to identify all human genes, making them accessible for further biological study.
2. History of Human Genome Project:
In U.S., the HGP was carried out by the Department of Energy (Human Genome Program) directed by Ari Patrinos, and National Institute of Health (Human Genome Research Institute) directed by Francis Collins. In 2001, Craig Venter, CEO of Celera Genomics, co-announced the completion (90%) of sequencing of the human genome (draft sequence).
The full sequence was completed and published in 2003 (finished sequence). More refined sequence is available in 2006 and correction of minor errors (1 less in 10000 DNA subunits) requires some time to come.
3. Aims of Human Genome Project:
The project was aimed for the benefits of humankind, generation of biologists and researchers have been provided with detailed DNA information that will be key to understand the structure, organization and function of DNA in chromosomes.
The project had several goals to achieve:
i. Identify all the approx. 25000-30000 genes in human DNA.
ii. Determine the sequences of the 3-billion chemical base pairs that make up human DNA.
iii. Store the information in databases.
iv. Improve tools for data analysis.
v. Transfer related technologies to the private sector.
vi. Address the ethical, legal and social issues that may arise from the project.
4. Human Genome Size:
A genome is an organism’s complete set of deoxyribonucleic acid (DNA), a chemical compound that contains the genetic instructions needed to develop and direct the activities of an organism. The human genome contains approx. Three billion base pairs which reside in 23 pairs of chromosomes.
Each chromosome contains hundreds and thousands of genes, and ranges in size from about 50000000 to 300000000 base pairs. The total number of genes is 30000 (approx.) and accounts for only 25% of the DNA; the rest is extra-genic DNA.
5. Human Genome Project Mapping:
Before beginning a sequencing project of the human genome, it was first necessary to produce a good framework map. Two general methods were developed for mapping human genome — standard method and whole genome short-gun method.
The standard method involves finding a segment of the genome and locating where it belongs. Genetic maps based on recombination frequencies between markers are useful in ordering genes. Molecular markers like RFLP, VNTRs (Microsatellites), STSs, SNPs have been used in mapping human genome.
The whole genome shotgun sequencing method involves shearing of genomic DNA followed by cloning, to produce a genomic library.
This is followed by sequencing of cloned DNA fragments at random, followed by shotgun assembly, i.e., the assembly of the fragment sequences into larger units on the basis of their overlaps. Groups of cloned DNA segments that can be aligned in an overlapping fashion to cover a region of the human genome are referred as contigs.
Yeast Artificial Chromosomes (YACs) were initially used as cloning agents when primary task was mapping. However, as the emphasis of the project shifted to sequencing. Bacterial Artificial Chromosomes (BACs) were used.
6. Human Genome Project Sequence:
Sequencing means determining the exact order of the base pairs in a segment of DNA. The primary method used by the HGP to produce the finished version of the human genetic code is map-based or BAC- based sequencing. The human DNA is fragmented into pieces that are relatively large, cloned in the bacteria, stored for replication as required.
A collection of BAG clones containing the entire human genome is called a BAC-library. In this method, each BAC clone is mapped to determine the location of that fragment in human chromosome and then the DNA letters are sequenced from each clone and their spatial relation to sequenced human DNA in other BAC clones.
For sequencing, each BAC clone is cut into still smaller fragments that are about 2000 bases in length. These pieces are called “sub-clones”. A “sequencing reaction” is carried out on these sub-clones. With the help of a computer then the short sequences are assembled into contiguous stretches of sequence of the clones.
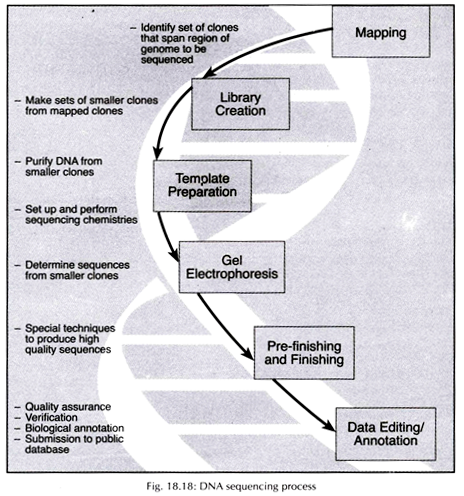
In a short the whole process can be summarized:
i. Chromosomes, which range in size from 50 million to 250 million bases, must first be broken into much shorter pieces (sub-cloning step).
ii. Each short piece is used as a template to generate a set of fragments that differ in length from each other by a single base that will be identified in a later step (template preparation and sequencing step).
iii. The fragments in a set are separated by gel electrophoresis (separation step).
iv. The final base at the end of each fragment is identified (base-calling step). This process recreates the original sequence of As, Ts, Cs and Gs for each short piece generated in the first step.
v. After the bases are ‘read’, computers are used to assemble the short sequences (in blocks of about 500 bases each called the read length) into long continuous stretches that are analysed for errors, gene coding regions, and other characteristics.
vi. Finished sequence is submitted to major public sequence databases, making Human Genome Project sequence data thus freely available to anyone around the world (Fig. 18.18).
The human genome reference sequence do not represent any one person’s genome. Rather the knowledge obtained is applicable to everyone because all humans share the same basic set of genes and genomic regulatory regions that control the development.
Researchers collected blood (female) or sperm (male) samples from different races like European, African, American (North, Central, South) and Asian ancestry and a few samples were processed as DNA resources.
7. Outcome of Human Genome Project:
i. The human genome contains 3164.7 million chemical nucleotide bases (A, C, T and G).
ii. The average gene consists of 3000 bases, but sizes vary greatly, largest known human gene is “dystrophin” – 2.4 million bases.
iii. Total number of genes estimated 30000 approx.
iv. Almost all (99.9%) nucleotide bases are exactly the same in all people.
v. 50% genes are unknown for function.
vi. Less than 2% genomes code for proteins.
vii. Repeated sequences (junk DNA) is 50% of the human genome. This may contribute to create new genes, to modify and reshuffle the existing genes.
viii. A-T rich regions are gene-poor and G-C rich regions are gene-dense. Chromosome-I has the most genes (2968) and the Y chromosome has the fewest (231).
ix. Scientists have identified about 1.4 million locations where single base DNA differences (SNPs) occur in human, these findings will help to localize the disease associated sequences in the chromosomes.
x. Finding the DNA sequences underlying such common diseases as cardiovascular disease, diabetes, arthritis and cancers is being aided by human variation maps (SNPs) generated in HGP.