In this article we will discuss about:- 1. Origin of Spontaneous Mutations 2. Evidence for Spontaneous Mutation 3. Random Nature 4. Mutation Rates.
Origin of Spontaneous Mutations:
Mutation involves changes in DNA. Several mechanisms are known that bring about alterations in DNA. These modifications may arise from error in DNA replication, damage to DNA from radiation. Errors occur during replication by substitution of frame shift in DNA sequence.
i. Substitution:
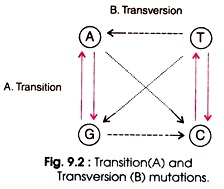
Substitution of base pairs is the most common mutation. During replication of DNA repair wrong base pairs are incorporated. Base pair substitutions are of two types, transition and transversion.
(a) Transition Mutation (Due to Tautomerism):
Transition is the replacement of one purine (e.g. A or G) by another purine (e.g. G or A) or one pyrimidine (e.g. T or C) by another pyrimidine (e.g. C or T). Four types of changes are possible in transition such as AT → GC, GC → AT, TA →CG, and CG → TA. (Fig. 9.2). During replication, errors in DNA arise with high frequency. Some of the bases exist in alternative forms having different base pairing properties.
Tautomerism is the relationship between two structural isomers that are in chemical equilibrium and readily change into one another. Generally the bases exist in keto form, but at a time they can take on either an imino or enol form (Fig. 9.3).
A rare form of adenine can pair with cytosine, and the end form of thymine can pair with guanine. These tautomeric shifts alter the hydrogen bonding characteristics of the bases, and permit for purine substitution or pyrimidine for pyrimidine substitution. These lead to stable modification in nucleotide sequences.
During replication an incorrect base is correctly hydrogen bonded and incorporated to the template strand. Even the editing system does not recognise it as incorrect. Later on when the base assumes its normal function, the mismatch repair system corrects the mismatching bases at this level.
However if the daughter strand containing the incorrect bases is methylated, the mismatch repair system fails to distinguish between the parental and daughter strands. Therefore, the incorrect bases exist in daughter strand and lead to mutation. Such mutation is known as transition mutation. Transition mutations are common, although most of them are repaired by various proofs reading function.
In a strand of DNA molecule a pyrimidine base C is substituted by the another pyrimidine base T which is the transition of base pair from one pyrimidine to another. After replication the error is retained in one strand and inherited in the progeny. The other strand remains normal. In the second generation after replication, purine base G is substituted by another purine base giving rise to mutant progeny.
(b) Transversion mutation:
Transversion mutation involves the substitution of purine by a pyrimidine or a pyrimidine by a purine. This type of mutation is rare due to steric problems of paring of purines with purines, and pyrimidines with pyrimidines. Eight types of changes are possible in transversion such as AT → TA, AT → GC, GC → CG, GC→ TA, AT → TA, TA → GC, CG → GC, and CG → AT (Fig. 9.2).
ii. Spontaneous Deamination of 5-Methyl-cytosine:
Another source of spontaneous mutation is a change in 5-methyl-cytosine (5-MeC), a methyl form of cystoine. Both MeC and C occasionally loose an amino group. The C is converted to U, and MeC to 5-MeU (thymine) (Fig. 9.4). The C pairs with A but not G, therefore, replication of a molecule containing GU base pair will finally result in substitution of an AT pair for the original GC pair.
The process in successive round of replication is GU→AU→AT. The uracil from DNA is removed by uracil glycosylase, hence the conversion of C→U rarely results in mutation. In addition 5-MeC loses an amino group and converted into 5- methyluracil which is actually the thymine.
There is no removal mechanism; hence GMeC pair becomes a GT pair which may be corrected by mismatch repair system. This system does not recognise MeC, therefore, it is present in a methylated strand. It is converted randomly sometimes correct GC pairs and sometimes wrong AT pair. Therefore, MeC acts as mutable site in a gene, and this site is called hot spot.
It should be noted that methylated bases are the normal constituents of DNA in some microorgamsm but not the mutagens. Many organisms contain both C and MeC. Methylation results in protection of DNA against enzymes synthesized by viruses. The bacteriophage T2 contains 5- hydroxyl-methyl-cytosine instead of cytosine.
iii. Frameshift Mutation:
If there is deletion or insertion of one or a few nucleotides in the DNA molecule, this shifts the reading frame of nucleotide sequences resulting in mutation. Therefore such mutation that results from shifting in reading frame backward or forward by one or more nucleotides is called frameshift mutation. (Fig. 9.5).
Generally this mutation occurs where there is a short repeated nucleotide sequence. Roth (1974) reviewed the frameshift mutation occurring in organisms. Fig. 9.5. shows an addition of one T due to slippage of the new strand (A), and a deletion (loss of two T base) occurs as a result of slippage of the parental strand (B). Hence, there is shifting in sequence of genetic code.
The same can be explained by an analogy of a code of words. If a gene gives the message: THE DOG CAN EAT JAM, each word of three letters represents a codon. After addition of a letter A after G, the new sentence becomes: THE DOG ACA NEA TJA M, which also is meaningless. Similarly, after deletion of C the new sentences become THE DOG ANC ATJ AM, which also is meaningless.
Evidence for Spontaneous Mutation:
Lederberg and Lederberg (1952) gave the direct evidence for the origin of spontaneous mutation in T1-r cells of E. coli without exposing to phage. This was presented by a procedure called replica plating (Fig. 9.1).
The steps of replica plating involve:
(i) Plating of bacteria on nutrient medium and proper incubation of plates for growth of bacterial colony,
(ii) Preparation of a solid support mounted with a piece of sterile velvet and gentle pressing it onto surface of plate supporting bacterial colonies i.e. the master plate,
(iii) Again gentle press of velvet (onto which some bacteria from each colony had sticked to fibres) on a Perti plate containing fresh medium that allowed to transfer the bacterial cells on fresh plate i.e. replica plate.
Precaution was taken to have the identical position of both the plates and velvet also. The master plate contained 107 colonies of bacteria growing on non-selective medium, whereas the replica plate contained T1 phage particles spread onto medium.
After proper incubation a few T1-r cells formed colonies in the same position on each of replica plate as in master plate. The master plate did not contain T1 phage. Therefore, appearance of T1-r cells on replica plate can be explained to occur due to the mutation for resistance that had taken place by chance in the master plate not exposed to T1 phage particles.
There is no means to know when and which cell will undergo mutation. Any gene of a cell of microorganisms is vulnerable to mutation. It is not sure, however, which gene will mutate. Every gene is subject to mutation, therefore, mutation occurs in a gene spontaneously and there is possibility of mutating the genes in a cell and more probability of occurrence of mutant allele in a given population of a microorganism.
In addition, it is not sure that mutation will be beneficial; it may be even detrimental too. Thus, the spontaneous mutation arises randomly in a population of organisms. Spontaneous mutations are rare ranging from 10-6 to 10-8 per generation depending on the gene and organism.
Random Nature of Mutation:
Before 1940’s it was believed that mutation occurs in bacterial population in response to a given selective condition i.e. a medium containing antibiotic substance. But Luria and Delbruck (1943) demonstrated the spontaneous and non-adaptive nature of mutation. These experimentations gave birth to microbial genetics.
They investigated the origin of mutation in E coli conferring resistance to phage T1 infection. The number of T1-r (r, resistant) mutant cells arising in different cultures of T1-s (s, sensitive) cells was compared with the number found in repeated samples of the same size taken from a single culture.
The result was analysed by a statistical test named as fluctuation test (Table. 9.1). Table 9.1 is based on the experiment in which twenty 0.2 ml culture and one 10 ml culture, each containing 103 cells/ml of T1-s bacteria were grown to 21 generation. The number of cells increased to 2.8 × 109 cells/ml.
Each of small culture and ten of 0.2 ml of large culture were plated onto individual plate which were already uniformly spread with 1010 particles of T1 phage (probably sufficient for destroying all T1-s cells). After incubation the number of T1-r colonies were counted.
The number of bacterial cells inoculated in each plate was the same (5.6 x 108) but the number of T1-r colonies depended on whether the cells had grown in small individual culture or in large culture. Out of 20 cultures no T1-r cell was detected in 11 small cultures.
In rest of 9 cultures the number of T1-r cells ranged from 1 to 107. On the other hand each of 10 samples of large culture had the same number. This experiment shows that T1-r cells arose by spontaneous mutation at different times in the growth of cultures in the absence of phage T1, therefore the number in different cultures varied greatly. However, if T1-r cells arise in response to phage, these should be about equal numbers in all population of the same size.
Table 9.1 : The number of T1 phage-resistant E.coli mutants in small individual cultures and in samples from a large bulk culture.
Mutation Rates:
The probability of a gene undergoing mutation in a single generation is known as mutation rate. For the study of population genetics, evolution and analysis of effect of environment mutagens and measurement of mutation rates are important.
In bacteria, a mutation can occur at any time during growth in culture. The mutant bacterial cell divides and increases the number at the same rate as the normal cells divide. Therefore, measuring the mutation rates is rather complicated.
Fluctuation test is an important method for estimation of mutation rates in bacteria. As given in column 2 of Table 9.1, a culture may contain many mutants, some mutants or none. If mutation rate per generation is µ, the probability of getting n mutants in a culture of N cells is Po, by the Poisson distribution- e-µN.
The total number of divisions of individual cell needed to yield N cells from 1 cell is N-1 where N is the large number of bacterial cells.
Thus, µN = In Po
or µ = (1/N) In Po …(1)
As shown in Table 9.1, 11 of the 20 small cultures did not contain T1-r cells. The average number of N cells per culture was 5.6 x 108.
Thus, the mutation rate can be calculated by putting the values in equation No. 1
µ = (1/5.6 X 108) In (11/20)
= 1.1 × 10-9 per cell per round of replication.
For the first time Benzer (1961) observed that at a particular site mutation occurs with high frequency than the other site within a gene. He mapped several thousand independently isolated rll mutation in T4 phage. Therefore, the favoured sites for mutation at high frequency were called hot spots. Coulondre (1978) have studied the molecular basis of base substitution hot spots in E. coli.