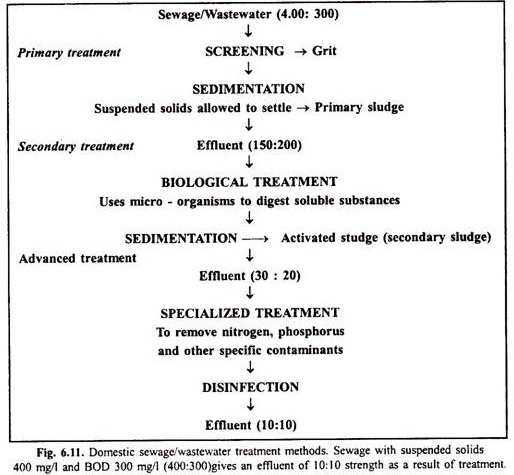
This article throws light upon the seven types of enzymes involved in DNA synthesis and cloning.
The seven types of enzymes are: (1) Alkaline Phosphatase (2) Terminal Transferase (3) Thermo-stable DNA Polymerases (4) Bacteriophage RNA Polymerases (5) Nucleases: DNase and RNase (6) Polynucleotide Kinase and (7) DNA Ligase.
Contents
Type # A. Alkaline Phosphatase:
Alkaline phosphatase removes 5′ phosphate groups from DNA and RNA (Fig. 13.4). It will also remove phosphates from nucleotides and proteins. These enzymes are most active at alkaline pH therefore known as alkaline phosphatase.
There are several sources of alkaline phosphatase that differ in how easily they can in-activated:
1. Bacterial alkaline phosphatase (BAP) is the most active of the enzymes, but also the most difficult to destroy at the end of the dephosphorylation reaction.
2. Calf intestinal alkaline phosphatase (CIP) is purified from bovine intestine. This is phosphatase most widely used in molecular biology labs because, although less active than BAP, it can be effectively destroyed by protease digestion or heat (75 °C for 10 minutes in the presence of 5 mM EDTA).
3. Shrimp alkaline phosphatase is derived from a cold-water shrimp and is promoted for being readily destroyed by heat (65°C for 15 minutes).
There are two primary uses for alkaline phosphatase in DNA manipulations:
1. Removing 5′ phosphates from plasmid and bacteriophage vectors that have been cut with a restriction enzyme. In subsequent ligation reactions, this treatment prevents self-ligation of the vector and thereby greatly facilitates ligation of other DNA fragments into the vector (e.g. sub-cloning).
2. Removing 5′ phosphates from fragments of DNA prior to labeling with radioactive phosphate. Polynucleotide kinase is much more effective in phosphorylating DNA if the 5′ phosphate has previously been removed.
It is usually recommended that dephosphorylation of DNAs with blunt or 5′-recessed ends be conducted using a higher concentration alkaline phosphatase or at higher temperatures than for DNAs with 5′ overhangs.
Type # B. Terminal Transferase:
Terminal transferase catalyzes the addition of nucleotides to the 3′ terminus of DNA. Interestingly, it works on single-stranded DNA, including 3′ overhangs of double-stranded DNA, and is thus an example of a DNA polymerase that does not require a primer. It can also add homo-polymers of ribo-nucleotides to the 3′ end of DNA. The much preferred substrate for this enzyme is protruding 3′ ends, but it will also, less efficiently, add nucleotides to blunt and 3′-recessed ends of DNA fragments. Cobalt is a necessary cofactor for activity of this enzyme.
Some of its uses are:
1. Labeling the 3′ ends of DNA:
Most commonly, the substrate for this reaction is a fragment of DNA generated by digestion with a restriction enzyme that leaves a 3′ overhang, but oligodeoxynucleotides can also be used. When such DNA is incubated with tagged nucleotides and terminal transferase, a string of the tagged nucleotides will be added to the 3′ overhang or to the 3′ end of the oligonucleotide (Fig. 13.5).
2. Adding Complementary Homopolymeric Tails to DNA:
This clever procedure was commonly used in the past to clone cDNAs into plasmid vectors, but has largely been replaced by other, much more efficient techniques. The principles of this technique are depicted in the Figure 13.6. Basically, terminal transferase is used to tail a linearized plasmid vector with G’s and the cDNA with C’s. When incubated together, the complementary G’s and C’s anneal to “insert” the cDNA into the vector, which is then transformed into E. coli.
Terminal transferase is a mammalian enzyme, expressed in lymphocytes. The enzyme purchased commercially is usually produced by expression of the bovine gene in E. coli.
Type # C. Thermo-stable DNA Polymerases (TAQ Polymerase):
It is interesting how some unimportant discoveries become something of immense practical importance after some time. Such is the history of Taq DNA polymerase. The original report of this enzyme, purified from the hot springs bacterium Thermus aquaticus, was published in 1976 (Fig. 13.7). Roughly 10 years later, the polymerase chain reaction was developed and shortly thereafter “Taq” became a household word in molecular biology circles. Currently, the world market for Taq polymerase is in the hundreds of millions of dollars each year.
The thermophilic DNA polymerases, like other DNA polymerases, catalyze template-directed synthesis of DNA from nucleotide triphosphates. A primer having a free 3′ hydroxyl is required to initiate synthesis and magnesium ion is necessary.
In general, they have maximal catalytic activity at 75° to 80°C, and substantially reduced activates at lower temperatures. At 37°C, Taq polymerase has only about 10% of its maximal activity. In addition to Taq DNA polymerase, several other thermostable DNA polymerases have been isolated and expressed from cloned genes. Three of the most used polymerases are described in the Table 13.2.
Type # D. Bacteriophage RNA Polymerases:
Phage-encoded DNA-dependent RNA polymerases are used for in vitro transcription to generate defined RNAs. Most commonly, the reaction utilizes ribo-nucleotides that are labeled with radio-nuclides or some other tag, and the resulting labeled RNA is used as a probe for hybridization. Other applications of in vitro transcription including making RNAs for in vitro translation or to study RNA struction and function. Several bacteriophage RNA polymerases are commercially available. They are named after the phage that encodes them, and either purified from phage-infected bacteria or produced as recombinant proteins.
Many of the plasmids used for carrying cloned DNA incorporate promoters for bacteriophage RNA polymerases adjacent to the cloning site. This allows one to readily obtain either mRNA sense or antisense transcripts from the inserted DNA. The process is often called run-off transcription, because the plasmid is cut with a restriction enzyme downstream of the inserted DNA, which causes the polymerase to fall off the template when it reaches that spot.
If we assume that the RNA transcribed (Fig. 13.8) has the polarity of a mRNA (e.g. sense), it is easy to modify the construct to express an antisense RNA – simply reverse the orientation of the transcribed region. Indeed, most plasmids used for in vitro transcription have two different phage polymerase promoters flanking the insertion site, which allows transcription of sense RNA with one polymerase and antisense with the other.
Type # E. Nucleases: DNase and RNase:
Most of the time nucleases are the enemy of the molecular biologist who is trying to preserve the integrity of RNA or DNA samples. However, deoxyribonucleases (DNases) and ribonucleases (RNases) have certain indispensible roles in molecular biology laboratories.
Numerous types of DNase and RNase have been isolated and characterized. They differ among other things in substrate specificity, cofactor requirements, and whether they cleave nucleic acids internally (endonucleases), chew in from the ends (exonucleases) or attack in both of these modes.
In many cases, the substrate specificity of a nuclease depends upon the concentration of enzyme used in the reaction, with high concentrations promoting less specific cleavages. The most widely used nucleases are DNase I and RNase A, both of which are purified from bovine pancreas: Deoxyribonuclease I cleaves double-stranded or single stranded DNA. Cleavage preferentially occurs adjacent to pyrimidine (C or T) residues, and the enzyme is therefore an endonuclease. Major products are 5′-phosphorylated di, tri and tetra nucleotides.
In the presence of magnesium ions, DNase I hydrolyzes each strand of duplex DNA independently, generating random cleavages. In the presence of manganese ions, the enzyme cleaves both strands of DNA at approximately the same site, producing blunt ends or fragments with 1-2 base overhangs. DNase I does not cleave RNA, but crude preparations of the enzyme are contaminated with RNase A; RNase-free DNase I is readily available.
Applications:
1. Eliminating DNA (e.g. plasmid) from preparations of RNA.
2. Analyzing DNA-protein interactions via DNase foot printing.
3. Nicking DNA prior to radio-labeling by nick translation.
Ribonuclease A is an endoribonuclease that cleaves single-stranded RNA at the 3′ end of pyrimidine residues. It degrades the RNA into 3′-phosphorylated mononucleotides and oligonucleotides. The major use of RNase A is eliminating or reducing RNA contamination in preparations of plasmid DNA.
Mapping mutations in DNA or RNA by mismatch cleavage:
RNase will cleave the RNA in RNA-DNA hybrids at sites of single base mismatches, and the cleavage products can be analyzed. A number of other nucleases that are used to manipulate DNA and RNA are described in the Table 13.3.
a. Exonucleases are enzymes (found as individual enzymes, or as parts of larger enzyme complexes) that cleave nucleotides one at a time from an end of a polynucleotide chain. These enzymes hydrolyze phosphodiester bonds from either the 3′ or 5′ terminus of a polynucleotide molecule (Fig. 13.9).
b. Endonucleases are enzymes that cleave the phosphodiester bond within a polynucleotide chain, in contrast to exonucleases, which cleave phosphodiester bonds at the end of a polynucleotide chain. Restriction endonucleases (Restriction Enzymes) cleave DNA at specific sites, and are divided into three categories, Type I, Type II, and Type III, according to their mechanism of action. These enzymes are often used in genetic engineering to make recombinant DNA for introduction into bacterial, plant, or animal cells (Fig. 13.10).
Type # F. Polynucleotide Kinase:
Polynucleotide kinase (PNK) is an enzyme that catalyzes the transfer of a phosphate from ATP to the 5′ end of either DNA or RNA. It is a product of the T4 bacteriophage, and commercial preparations are usually products of the cloned phage gene expressed in E. coli.
The enzymatic activity of PNK is utilized in two types of reactions:
1, In the “forward reaction”, PNK transfers the gamma phosphate from ATP to the 5′ end of a polynucleotide (DNA or RNA). The target nucleotide is lacking a 5′ phosphate either because it has been dephosphorylated or has been synthesized chemically.
2. In the “exchange reaction”, target DNA or RNA that has a 5′ phosphate is incubated with an excess of ADP, PNK will first transfer the phosphate from the nucleic acid onto an ADP, forming ATP and leaving a dephosphorylated target. PNK will then perform a forward reaction and transfer a phosphate from ATP onto the target nucleic acid (Fig. 13.11).
As you might expect, the efficiency of phosphorylating via the exchange reaction is considerably less than for the forward reaction. In addition to its phosphorylating activity, PNK also has 3′ phosphatase activity, although this has little significance to molecular technologists.
There are two major indications for phosphorylating nucleic acids and hence uses of PNK are:
1. Phosphorylating linkers and adaptors (fragments of DNA ready for ligation) which require a 5′ phosphate. This includes products of polymerase chain reaction, which are typically generated using non-phosphorylated primers.
2. Radiolabelling oligonucleotides, usually with 32P, for use as hybridization probes. PNK is inhibited by small amounts of ammonium ions, so ammonium acetate should not be used to precipitate nucleic acids prior to phosphorylation. Low concentrations of phosphate ions, or NaCl concentrations greater than about 50 mM, also inhibit this enzyme.
Type # G. DNA Ligase:
The term recombinant DNA includes the concept of recombining fragments of DNA from different sources into a new and useful DNA molecule. Joining linear DNA fragments together with covalent bonds is called ligation. More specifically, DNA ligation involves creating a phosphodiester bond between the 3′ hydroxyl of one nucleotide and the 5′ phosphate of another.
The enzyme used to ligate DNA fragments is T4 DNA ligase, which originates from the T4 bacteriophage. This enzyme will ligate DNA fragments having overhanging, cohesive ends that are annealed together, as in the EcoRI (Fig. 13.12). This is equivalent to repairing “nicks” in duplex DNA. T4 DNA ligase will also ligate fragments with blunt ends, although higher concentrations of the enzyme are usually recommended for this purpose.