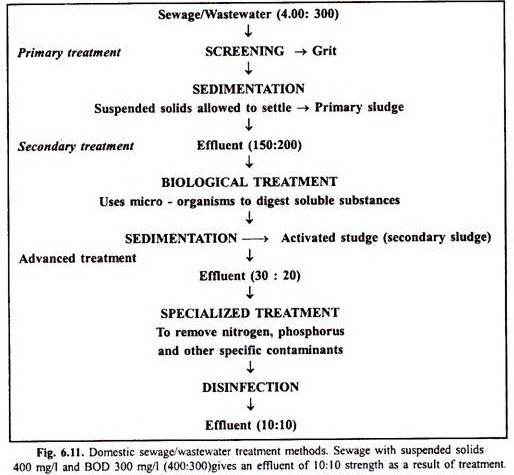
In this article we will discuss about the structure of herpesviruses (explained with diagram).
All of them are morphologically identical. They are large with a varying diameter from 140-170 nm. The nucleocapsid is spherical icosahedral and of about 100 nm diameter which is enclosed in a 30 nm thick glycoprotein lipid envelope.
Thickness of the envelope indicates for the presence of a matrix. The nucleocapsid contains the spikes (Fig. 17.10). The structure of the herpesvirus particle is very complex. Viral envelope that surrounds the nucleocapsid is derived from the inner nuclear membrane of the host cell. Capsid of the herpes virus is an icosahedron of triangulation number T = 16.
The capsid measures about 100 nm which is made up of hollow columnar capsomers (162) forming an icosahedral symmetry. There are a total of 150 hexamers and 12 pentamers. Outside the capsid is the tegument (matrix), a protein-filled region which appears amorphous in electron micrographs.
The envelope contains numerous viruses-encoded glycoproteins that are incorporated into the envelope. The glycoproteins are visible as ‘spikes’ that project from its surface (Fig. 17.10). When the envelope breaks and collapses away from the capsid, negatively stained virions are released and have a typical ‘fried-egg’ appearance.
The nucleic acid is a linear dsDNA which is converted into a circular DNA molecule upon a slight digestion either 3′ or 5′ end. It has also been observed that the DNA consists of two fragements (i.e. one long and the other short DNA molecule) which are covalently linked at the identical region, but they replicate independently or may be the recombination product of different orientation.
Genomes of the herpesvirus contain multiple repeated sequences. Depending on the number of these sequences, genome size of various isolates of a particular virus can vary by about 10 kb sequences. Herpesviruses have a large genome consisting of about 235 kb dsDNA.
All herpesvirus genomes have a unique long (UL) and a unique short (US) region bounded by inverted repeats (see arrows) (Fig. 17.11). The repeats allow rearrangements of the unique regions and herpesvirus genomes exist as a mixture of four isomers. Complex viruses contain about 35 virion proteins.
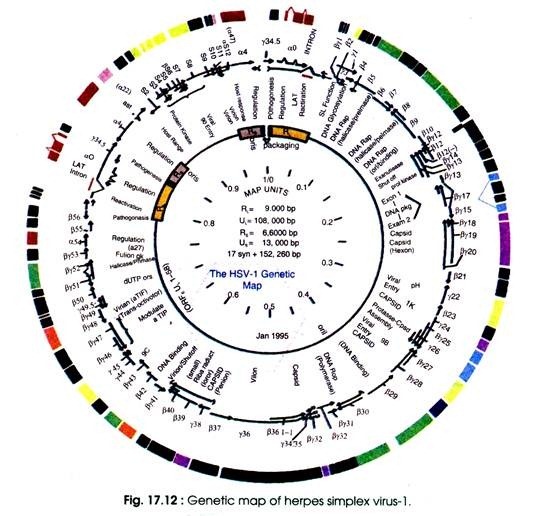
Genome of herpes simplex virus-1 has been sequenced and its genetic map has also been prepared (Fig. 17.12).