In this article we will discuss about the replication of herpesviruses.
Sequential interaction takes place between specific viral membrane glycoproteins and cell membrane receptors (e.g. cell surface proteoglycans which are heparin sulphates). On the basis of complementary residues present on host cell and viral membrane, binding between virus and host cell membrane takes place before entering the virus within the host cell.
Much of the recent work elucidating these receptors and the viral glycoproteins interacting with them has been carried out in the laboratories of P. Spear (Northwestern University), and G. Cohen and R. Eisenberg of the University of Pennsylvania.
Consequently, fusion between host and virus membranes occurs which leads to penetration of viral capsids inside the host cell. After entering the host cell, nucleocapsid is transported to the nuclear pores, where viral DNA is released into the nucleus.
The viral genome is accompanied by the a-TIF protein which functions in enhancing the immediate early viral transcription via cellular transcription factors. The virion-associated host shutoff protein (vhs-UL 41) appears to remain in the cytoplasm where it causes disaggregation of polyribosomes and degradation of cellular and viral RNA (Fig. 17.13).
After infecting a host cell, the HSV has to decide quickly, whether to proceed for ‘productive (vegetative) infection’ or to establish a ‘latent infection’. When viral DNA migrates to nuclear pods it is either circularized by cellular DNA repair enzymes acting on the ‘A’ sequences or remains linear through the action of the immediate-early ICPO protein which inhibits cellular DNA repair.
The latent infection ensues in the former case. While in the latter, the productive replication cycle of HSV gene expression characterized by a progressive cascade of increasing complexity takes place.
The earliest genes expressed in the ‘immediate-early’ are important in ‘priming’ the cell for further viral gene expression and in mobilizing cellular transcriptional machinery.
This phase is followed by the expression of a number of genes either involved directly or indirectly in viral genome replication. Finally, after genome replication viral structural proteins are expressed in high abundance during the ‘late phase’ (Tables 17.4).
The replicative cycle of HSV is rapid as compared to many other herpesviruses, but this general temporal classification fits the productive replication cycle of all as well as smaller nuclear replicating DNA viruses such as adenoviruses and papovaviruses.
(a) Expression of Immediate Early Genes:
There are five HSV genes viz., α-4-lCP4, α-0-ICP0, α-27-ICP27/UL 54, α-22-ICP22/US1 and α-47-ICP47/US12 which get expressed and functions at the earliest stages of the productive infection cycle. This stage of infection is termed as the ‘immediate-early’ stage. This stage is mediated by the action of virion-associated α-TIF through its interaction with cellular transcription factors at specific enhancer elements that are associated with the individual a-transcript promoters.
The a-TlF binds to cellular Oct-1, which has bound to TAATGARAT sequences in enhancers upstream of the immediate early promoters. This results in efficient assembly of the pre-initiation complex at the TATA box and pol II-mediated transcription. Immediate-early transcripts are transported to the cytoplasm, translated, and the immediate-early proteins migrate back to the nucleus.
All further transcription requires the action of these proteins, especially the α-4 protein. It acts as a generalized transcription activator and works by binding with multiple sites on the genome and then interacts with nearby TATA boxes to facilitate assembly of pre-initiation complexes. At the level of transcription or mRNA expression, proteins encoded by the α-4, α-0, and α-27 transcripts activate viral gene expression.
They interact to form nuclear complexes with viral genomes. There are two other proteins, α-22 and α-47, which is dispensable for virus replication in many types of cultured cells but α-22, is required for HSV replication.
In other viruses it plays a role to maintain virus ability to replicate in a broad range of cells in the host. The α-47 protein modulates host response to infection by interfering through presentation of viral antigens on the surface of infected cells.
(b) Expression of Delayed Early Genes:
Activation of transcriptional machinery of the host cells by the action of a gene products results in the expression of the early or β-genes. The thymidine kinase transcript is one of the promoters for such genes which serve as the models for typical eukaryotic promoters.
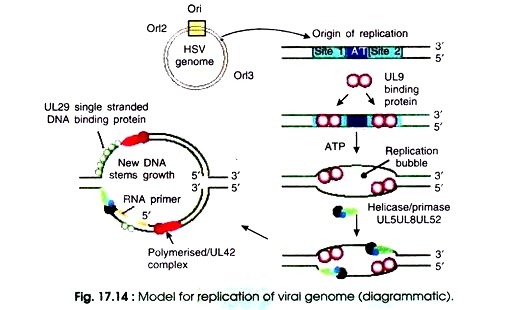
Of these there are seven proteins which are necessary and sufficient for replication of viral DNA under all conditions such as DNA polymerase (UL30), DNA binding proteins (UL 42 and UL 29 or ICP8), ori binding protein (UL 9), and the helicase/primase complex (UL 5, UL 8, and UL 52). When sufficient levels of these proteins have accumulated within the infected cell, there starts the replication of viral DNA (Fig. 17.14).
The other early proteins are involved in increasing the amount of DNA pool in the infected cells, whereas the proteins act as repair enzymes for the newly synthesized viral genomes.
These accessory proteins are non-essential for replication of virus where the cellular products can substitute for their function in one or another cell types. After disrupting such genes a profound effect is caused on viral pathogenesis, and/or its ability to replicate in specific cells.
(c) Genome Replication and Expression of Late Genes:
Replication of viral DNA is the main event in the viral replication cycle. High levels of DNA replication results in faster cell destruction. DNA replication also significantly influences the gene expression of viruses. Following the start of DNA replication, early expression is significantly reduced or shut off, whereas the late genes begin to be expressed at high levels.
Transcripts expressing late genes can be divided into two subclasses: leaky-late and strict-late. These differ in the amount of transcription observed prior to genome replication. Promoters controlling expression of both classes are similar in that both have elements near the transcription start site (cap site) which are required for promoter activity, but the location of other elements can differ.
The leaky-late transcripts are expressed at low levels before start of DNA replication but reach maximum expression after the initiation of replication of viral DNA.
In contrast, the strict late transcripts are difficult to detect at all until viral DNA replication has been started. These are controlled by promoters that have different functional architectures, but both have sequence elements downstream of the TATA box important in stabilizing the formation of pre-initiation complexes.