The below mentioned article provides a study note on the Operon Concept.
The DNA present in a cell contains genetic information for all the proteins required by an organism and may contain from few to thousands of genes. All the functional genes do not make polypeptides all the times. Though the different types of cells in the body of an organism differ in their structure and function, their genes are identical.
As development proceeds the activity of some genes increases while those of others decreases or stops, i. e., the genes are switched on and off at different times.
This process is referred to as differential gene action. When the genes are active they direct the synthesis of enzymes and proteins which affect certain phenotypic traits. The metabolic products may influence the synthesis of enzymes (feed-back or inhibition of end product).
Although the cell may contain many genes which produce as many enzymes as their number, only the enzymes required at a particular time are produced. Therefore, the flow of information from DNA (gene) to protein is regulated in some way or the other.
The mechanism of regulation of gene action has been studied extensively in certain micro-organisms such as Escherichia coli and some higher organisms. A hypothesis to explain induction and repression of enzyme synthesis was first put forward by Francois Jacob and Jacques Monod (1961). For this and other brilliant contributions they were awarded Nobel Prize in Medicine in 1965.
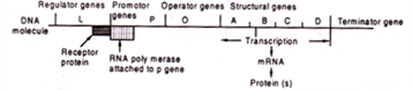
The scheme proposed by Jacob and Monod is called Operon Model which consists of the following components (Fig. 20.3). Operon is a group of structural genes whose transcription is regulated by action of regulator gene, promotor gene, operator gene and terminator gene.
Recent studies in microbial genetics have clearly indicated that not all the genes act by determining the structure of given proteins or enzymes. Some of them regulate the action of other genes and are called regulator genes. Regulation may be brought about either by suppression of protein synthesis or by induction of the process.
Structural Gene:
These are directly concerned with the synthesis of cellular proteins. The structural genes produce «RN As through transcription and determine the sequence of amino acids in the synthesized proteins. As there are many proteins in the cells, there are as many structural genes under common control.
Each structural gene may be controlled independently and that transcribes a separate mRNA molecule or all the structural genes under an operon may form one long polycistronic or polygenic mRNA molecule.
Eukaryotes have only a single functional site for producing monogenic mRNA, whereas, in prokaryotes polygenic mRNAs are common. A small segment of DNA molecule in bacterium may have several structural genes which may transcribe one long polycistronic mRNA molecules. This controls the synthesis of many proteins.
Operator Gene:
Operator gene is located adjacent to first structural gene. It determines whether the structural gene is to be repressed by the repressor protein, a product of regulator gene. The operator gene is recognised by repressor protein which binds to the operator forming an operator-repressor complex.
The operator genes as well as the group of genes that are under it are known as operons. The operator genes are carried in the chromosome and show recombination also.
Promoter Gene:
It is continuous with the operator gene and is located just before operator. The complete nucleotide sequence of control region in some bacteria extends from the termination codon (ACT on DNA or UGA on mRNA) of the structural gene and induces the promoter and operator genes. The promoter region according to Pribnow (1975) has three essential elements which are constant in position with respect to each other.
These elements are:
(i) A recognition sequence,
(ii) A RNA-polymerase binding sequence, and
(iii) An mRNA initiation site.
The recognition site is located outside the polymerase binding site, i.e., the region of DNA protected from the action of DNAse. RNA polymerase first binds to DNA by forming a complex with the recognition sequence. Then it binds with the binding sequence to produce the pre-initiation complex.
The binding site consists of a group of 7 bases present in a constant position in the protected fragments. RNA initiation site starts the mRNA transcription on one or two bases near the binding sequence. The starting sequence is located in the DNA fragment protected from DNAse action.
Regulator Gene:
These genes direct the synthesis of protein which may be either an active repressor or an inactive repressor (apo-repressor). Repressor protein has one active site for operator recognition and the other active site for inducer. When inducer binds with repressor, it distorts the molecule so that operator site no longer recognizes the operator gene.
Active repressor protein has an affinity for the operator gene. In absence of an inducer protein the repressor binds to the operator gene and blocks the path of RNA polymerase.
Thus the structural genes are unable to transcribe mRNA and consequently protein synthesis does not occur. In presence of an inducer the repressor protein binds to the inducer to form an Inducer-repressor complex or co-repressor.
The repressor when linked to an inducer undergoes a change which makes it ineffective and as a result it cannot bind to the operator gene and the protein synthesis is possible.
Effector or Inducer:
An inducer or effector is a small molecule—a sugar, an amino acid or a nucleotide that can be linked to a regulator or repressor protein to change its ability to interact with an operator or promoter gene.
Jacob and Monod have suggested that a regulator protein will bind to an operator and inhibit operon expression as long as the effector or inducer is absent, but if the effector molecule appears in the intracellular environment it will bind to the regulator protein and thereby pull the regulator away from the operator.
According to Jacob and Monod concept, operon is a group of contiguous structural genes showing coordinate expression and closely associated sites. This definition does not presume any particular mechanism of regulation, negative or positive.
In the case of negative control of operon expression of the controlling elements are so coded that transcription of the operon occurs in the absence of any cytoplasmic regulatory component (repressor) specific for the particular system.
In the case of positive control system the coding at the initiator controlling site prevents the expression of operon in the absence of any specific regulatory product. The operon represents a unit of polarized coordinated genetic transcription in prokaryotes that is initiated and regulated at the operator site.
It consists of the promoter, the operator, one or more structural genes coding for enzymes performing related metabolic functions, and the terminator (Fig. 21.4). Transcription of the operon which eventually results in a polycistronic mRNA starts at the promoter where the RNA-Polymerase binds to DNA. Synthesis of mRNA starts if the operator is free of repressor protein (negative control).
In the repressible system repressor protein formed by regulator gene is inactive, therefore, it does not block the operator and protein synthesis takes place. Repressor is activated in the presence of co-repressor and the repressor-co-repressor complex makes the operator gene inactive. As a result of this, the protein synthesis cannot take place.
Regulator genes produce macromolecules that act upon the operator genes to turn it on or off. A single regulator gene can affect the synthesis of different proteins whereas the structural gene obeys the one gene-one enzyme rule.
If the repressor substances produced by regulator genes act upon operator genes, they depress their activity and then the latter do not permit the structural genes to produce their mRNAs. This is said to be the off condition (Fig. 21.5).
Regulator genes sometime produce inducer substances. If this substance is present in the nucleoplasm, it combines with the repressor substance, these two repressor and inducer substances do not act upon operator genes (O), they activate the operator genes and allow operator genes to turn on the structural genes as shown below in Fig. 21.6.
Transcription terminates at the end of operon defined by the terminator site where the mRNA is released. The rate of transcription in a given operon is a function of two parameters.
(i) The frequency of transcription initiation, and
(ii) The rate of transcription.
Some operons appear to have extra regulator loci in addition to the usual promoter site adjacent to operator, and extra promoter between structural genes has been found. Since extra promoter gene is not linked to the operator, some transcription of the last few genes occurs even when the system is switched off.