The Genetic Code:
The four letter language of nucleic acids is translated into the twenty letter language of the proteins viz the genetic code. The early genetic experiments indicated that each amino acid is coded for by a certain number of successive nucleotides in DNA. The best way of deciphering the genetic code therefore would be to compare the nucleotide sequence of a gene containing segment of DNA with the amino acid sequence of its specific protein.
This could not be achieved until the early 1960s due to lack of knowledge about the existence of mRNA. Once it became established that mRNA carries information from DNA to a specific protein, the problem was simplified. Thereafter the study of the genetic code was approached biochemically, and consisted in analysing the relationship between the nucleotide sequence of mRNA and the amino acid sequence of its protein.
Gradually the code words for all the amino acids were discovered and confirmed by genetic and biochemical evidence. The elucidation of the genetic code is one of the greatest scientific achievements in recent times.
The Triplet Code:
The 4 code letters of DNA specifying the sequence of 20 amino acids are the 4 bases A, T, G and C. If the 4 bases are arranged in groups of 2, it is possible to have 42 = 16 pairs or combinations of code words. Since 16 pairs are not enough for the 20 amino acids, the code for each amino acid must contain more than 2 bases. The 4 bases taken 3 at a time can specify 43 = 64 different amino acids. The genetic code for amino acids therefore, consists of triplets of bases.
Identifying Code Words for Amino Acids:
Nirenberg and Matthaei in 1961 devised a cell free system for protein synthesis. They could break open cells and utilise a mixture of the cell components for synthesizing proteins. When radioactively labelled amino acids were supplied to the cell extracts, they were detected in the newly synthesised proteins.
Nirenberg and Matthaei also used artificially synthesised mRNA in the cell free system. When they added a synthetic mRNA consisting only of one base uracil (Poly U), the protein synthesised consisted only of one amino acid phenylalanine. Since U codes for polyphenylalanine, it suggests that the triplet UUU codes for the amino acid phenylalanine.
By similar experiments it was found out that the synthetic polyribonucleotide poly C codes for poly-proline, that is, the triplet CCC codes for the amino acid proline. The synthesis of polyribonucleotide (mRNA) such as poly U in the laboratory was possible due to the earlier discovery of the enzyme polynucleotide phosphorylase.
This enzyme was first isolated by Ochoa (Nobel Laureate) and his colleagues from Azobacter vinelandii. Ochoa and his colleagues also devised a cell free system that could be used for protein synthesis. They found out that the triplet AAA codes for lysine.
Attempts were now made to synthesise random RNA copolymers in cell free systems using 5′-ribonucleoside diphosphates and the enzyme polynucleotide phosphorylase.
In this method the enzyme catalyses the addition of the 5′-ribonuclease diphosphates to the 3′ ends; the availability of the diphosphates depending on their concentration. That is, if the concentration of ADP is 5 times that of CDP, then the copolymer synthesised will also contain 5 times the number of Cs as compared to As, but in random order.
The coding triplets in such a copolymer lead to the synthesis of a polypeptide chain containing mainly lysine, threonine and proline. That is because C and A can form various possible triplets such as CCA, CAC, AAC, ACA, CCC, AAA. As the proportion of as is five times more than Cs in the copolymer, the frequency of AAA, AAC, and CAA codons would be correspondingly higher than the remaining codons.
By use of synthetic copolymers most of the possible codons could be assigned to amino acids. However, this technique could only determine the composition of the code words. The sequence of nucleotides in the triplets that is, the spelling of a codon still remained unknown.
The Triplet Binding Assay:
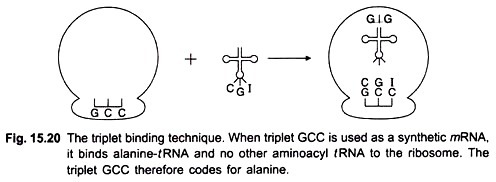
In 1964 Nirenberg and Leder discovered a direct method for determining the sequence of nucleotides in the code. They found that synthetic homopolymers such as poly U stimulated the binding of its specific aminoacyl-tRNA (i.e. Phenylalanine-tRNAphe) to ribosomes; simultaneously these ribosomes bind to the synthetic mRNA (poly U).
No other aminoacyl-/RNA will bind to ribosomes and to poly U. Similarly when the synthetic mRNA consisted of poly A and ribosomes were added, it induced the binding of lysine-tRNA and not of any other aminoacyl-tRNA. This is called triplet binding assay or tRNA-binding technique. Later on this test was also applied to polyribonucleotide copolymers used as synthetic mRNA (Fig. 15.20).
Nirenberg and Leder also found that triplet code had a polarity. Thus the triplet GUU stimulates the binding of valine tRNAval, whereas UUG binds leucine tRNALeu. They next proceeded to synthesise trinucleotides of known base sequence.
By the triplet binding method they could determine which aminoacyl-tRNA was specifically bound to ribosomes in the presence of a trinucleotide of known sequence. In this way the sequence of nucleotides in a triplet was established for many of the amino acids.
H.G. Khorana and his colleagues made some very outstanding contributions in this field. Khorana was recipient of the Nobel Prize in 1968. They provided experimental evidence for codon sequences using synthetic mRNAs of known nucleotide sequences.
They synthesised various copolymers with two, three and four kinds of bases. For example, when copolymer consisting of poly GUA was prepared, it had the sequence GUAGUAGUAGUAGUAGUA . First they synthesised two complementary deoxyribonucleotides each containing nine residues that is d(TAC)3 and d(GTA)3.
Then they used these two oligonucleotide chains as templates on which long DNA chains were synthesized from the four deoxyribonucleoside triphosphates by the enzyme DNA polymerase I. Neither of the two oligonucleotide chains when used alone could serve as templates. But when both chains were present, d(TAC)3 acted as template for synthesis of poly d(GTA), and d(GTA)3 served as template for synthesis of poly d(TAC). These two long complementary chains formed a double helical structure.
After this Khorana and colleagues prepared long polyribonucleotide chains with a sequence corresponding to poly d(TAC) and poly d(GTA). This was achieved by using the poly d(TAC) and poly d(GTA) double helix as template for synthesis by RNA polymerase. The choice as to which DNA strand would be transcribed by RNA polymerase depended upon the ribonucleoside triphosphates.
When GTP, UTP and ATP were added, the polyribonucleotide synthesised from the poly d(TAC) template strand was poly GUA. The other strand was not transcribed due to absence of CTP from incubation mixture. But when CTP, UTP and ATP were supplied, poly UAC was synthesised from the second template strand. By this method two long poly-ribonucleotides were synthesised each having defined repeating sequences.
By 1965 the work of Khorana, Nirenberg and Mathaei, and of Ochoa had resulted in identification of the coding triplets for all the amino acids (Table I by Crick).
Features of the Genetic Code:
1. The code is commaless, that is there is no nucleotide reserved for punctuations in order to indicate the end of one codon and the beginning of the next. The triplets are read successively one after the other.
2. It is a degenerate code in the sense that most of the amino acids are coded for by more than one triplet. For example, the amino acids tyrosine, histidine, glutamic acid and some others are coded by two triplets each; and amino acids arginine, serine and leucine are each coded by 6 triplets. There are only 2 amino acids tryptophan and methionine which are coded by a single triplet.
3. Out of the 64 triplets, 3 do not code for any amino acid. These are, UAG, UAA and UGA. These triplets serve as signals for the termination of polypeptide chains.
4. The first two bases in the triplet specify the amino acid; the third base is less specific.
According to Crick (1966) the third base tends to “wobble” on the basis of which he proposed his wobble hypothesis. Crick noticed that the degeneracy of the code involves only the third base in the codon in most cases. Thus UUU and UUC both code for phenylalanine; UUA and UUG for leucine; and GCU, GCC, GCA and GCG for alanine.
Crick explained the wobble hypothesis on the basis of codon-anticodon base pairing. The pairing of mRNA codon and tRNA anticodon is antiparallel. Conventionally the nucleoside at the 5′ end is written to the left. Therefore the third position of the codon is the first position of the anticodon.
This leads to the following type of pairing:
Crick proposed these pairing rules (wobble rules) for the codon position. According to these rules it is possible to relate more than one species of tRNA each with a specific anticodon, to the reading of specific codons.
Thus the codons for glutamine CAA and CAG would be expected to pair with a single anticodon UUG. However, the hydrogen bonds between the bases of the codon and anticodon are loose enough to allow the complex to dissociate; this is required during active protein synthesis.
5. The genetic code is universal: It is applicable to tobacco mosaic virus, bacteriophages, E. coli, animals and man. In all the species tested the code triplets are identical.
Genetic Evidence for the Code:
Comparative studies of mutations (changes in gene structure) and corresponding alterations in amino acid sequence of a specific protein have confirmed the validity of the genetic code. Some of these studies relate to the effects of mutation on the amino acid sequence of haemoglobin, the protein present in red blood corpuscles.
A molecule of haemoglobin contains 4 polypeptide chains, two identical α chains consisting each of 141 amino acids, and two β chains, also identical and containing 146 amino acids in each. In 1956 Ingram investigated the abnormal haemoglobin (S) from patients having sickle cell anaemia.
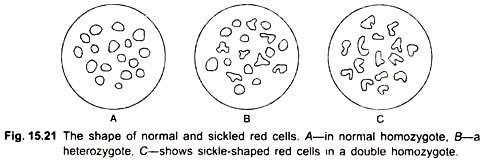
He found that in the β chain, the amino acid in the sixth position is valine, whereas in normal hemoglobin (A) it is glutamic acid. There is already a valine in the first position. The two valines in positions 1 and 6 become associated and presumably lead to a conformation that causes the erythrocyte to become sickle-shaped (Fig. 15.21).
That hemoglobin S results from mutation in the gene can be observed from the inheritance of the protein. Individuals with sickle-cell anemia are homozygous for the gene controlling haemoglobin S. When such a person marries a normal person, all their children are heterozygotes as they carry one gene for haemoglobin S and one gene for normal haemoglobin A.
The red blood cell of heterozygous individuals contain roughly half normal haemoglobin and half sickle-cell haemoglobin. Such persons are said to be carriers of the sickle-cell trait and lead almost normal lives. Most homozygotes die of sickle cell anaemia before the age of 30. Under conditions of low oxygen, sickle cells tend to clump together. The clumps block the capillaries so that blood supply to some regions is affected.
Many other kinds of mutant haemoglobins with one abnormal amino acid in the β chain, some in the α chain have been investigated. The substitution of the amino acid is explained on the basis that only a single nucleotide in the coding triplet is altered. The changed triplet becomes a codon for another amino acid.
Single point mutations have been studied for their effect on the sequence of viral coat proteins. The tobacco mosaic virus consists only of RNA and protein Mutations were induced in viral RNA by treatment with nitrous acid before infecting tobacco leaves.
The lesions produced by mutant strains of virus are different from those of the normal virus. The proteins of the mutant strains were isolated by Wittmann. He could analyse 29 different amino acid substitutions in the polypeptide chains which consisted of 157 amino acids.
The genetic code has also been confirmed from the study of frameshift mutations, so called because the normal reading frame of nucleotide triplets becomes changed due to addition or deletion of a single base. Frameshift mutations produce defective proteins with altered amino acid sequence in corresponding positions thus verifying the genetic code.
Crick, Brenner and their colleagues in 1961 conducted genetic experiments with such mutations and established the following:
(a) Triplets of bases code for each amino acid;
(b) The code is non-overlapping;
(c) The sequence of bases is read from a fixed starting point, the first nucleotide of the sequence; if the starting point is displaced by one base, the reading of the subsequent triplets becomes incorrect;
(d) The code is degenerate.
Crick and his colleagues experimented with proflavine-induced mutations in rII mutants of bacteriophage T4. Proflavine is an acridme mutagen that either adds or deletes one or more bases in DNA. The wild type TA grows on both E. coli strain B and on E. coli K12 (λ). The rII mutants of T4 do not grow on strain K, and produce r type plaques on E. coli B. Crick et al., (1961) worked with a mutant P 13 renamed FCO in the B cistron of rII region.
Crick argued that if a mutation is due to addition of a base, then deletion of a base could revert the mutant to the wild type. They found that reversion was actually due to a second mutation involving a suppressor in the same gene. The wild type so produced was a double mutant. They could map 18 suppressors in the B region of mutant FCO.
The suppressors produce an r mutation; the phages do not grow on E. coli K and produce r plaques on B strain. The point to note was that although the rII mutants looked alike, each had a structural defect in a different region of the B cistron.
The experiments of Brenner had already shown, that by double infection of E. coli the defects in genes of two viruses can be recombined to yield progeny having both defects in one gene. The conclusion that a nucleotide had been added or deleted in a mutant was arrived at in the following way.
The viruses were divided into two groups. When the defects in any two viruses of the same group were recombined, the resulting rII mutants would not grow on E. coli K. But if defects in viruses belonging to the 2 groups were combined, the resulting progeny viruses would grow on strain K and produced wild type plaques on E. coli B.
Crick and his colleagues assumed and later proved by further experiments that the 3 nucleotides of a single triplet or codon are determined solely by their position relative to the first nucleotide of the gene.
Suppose for the sake of simplicity that the rII mutants form r plaques due to a sequence of triplets repeated as follows:
CATCATCATCATCAT……
Suppose that one rll mutant had a deletion of the fourth base in the sequence and that another mutant got an extra base G inserted between the 6th and 7th bases, the sequence would then be
These triplets code for different amino acids and the resulting protein produces r type plaques. If by double infection bases 1 to 5 of the first mutant virus were combined with bases 6 onwards of the second, the resulting virus would have DNA of original length and sequence
C A T A T G C A T C A T C A T C A T ….
Thus after the second change the triplets C A T are restored. Such a sequence could produce a protein with one incorrect amino acid (coded by ATG). It may be assumed that if the deletion and insertion are not too far apart, they could produce a protein normal enough to enable the virus to grow on E. coli K. Indeed, by applying the mapping technique of Benzer, and increasing the distance between the deletion and the insertion, this assumption was found to be true.
Un-Coded Amino Acids in Proteins:
Some proteins contain amino acids that are not coded for by a triplet of nucleotides. For example, hydroxyproline and hydroxylysine in collagen, desmosine in elastin, N-methyl-lysine in some muscle proteins, and a few others. Such amino acids arise due to post-translational modification of amino acids that have been inserted into the polypeptide after instructions from their specific triplets.