This article throws light upon the top five screening strategies of gene libraries.
The top five screening strategies are: (1) Screening by DNA Hybridization (2) Screening by Colony Hybridization (3) Screening by PCR (4) Screening by Immunological Assay and (5) Screening by Protein Function.
Contents
1. Screening by DNA Hybridization:
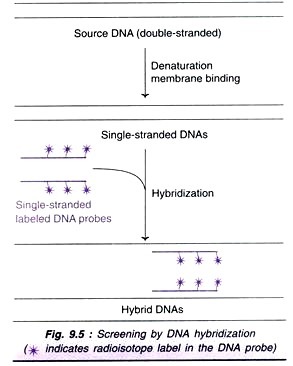
The target sequence in a DNA can be determined with a DNA probe (Fig. 9.5). To start with, the double-stranded DNA of interest is converted into single strands by heat or alkali (denaturation). The two DNA strands are kept apart by binding to solid matrix such as nitrocellulose or nylon membrane.
Now, the single strands of DNA probe (100-1,000 bp) labeled with radioisotope are added. Hybridization (i.e., base pairing) occurs between the complementary nucleotide sequences of the target DNA and the probe. For a stable base pairing, at least 80% of the bases in the two strands (target DNA and the probe) should be matching. The hybridized DNA can be detected by autoradiography.
DNA Probes:
The DNA probes used for screening purpose can be synthesized in many ways.
Random primer method:
Radioisotope labeled DNA primers can be produced by this technique (Fig. 9.6). The double- stranded DNA containing the sequence needed to serve as a probe is denatured. A mixture of synthetic oligonucleotides, with all possible combinations of bases (A, G, C and T), with a length of 6 nucleotides each serve as primers. Some of these primers with complementary sequences will hybridize with the template DNA. This occurrence is entirely by chance and the probability is reasonably good.
By the addition of four deoxyribonucleotides (one of them is radiolabeled) and in the presence of the enzyme DNA polymerase of E. coli (Klenow fragment), the primers are extended on the template DNA. Since a radioactive label is used, the newly synthesized DNA fragments are labeled at appropriate places, and these are the DNA probes. A number of labeled DNA probes can be produced from an unlabeled template DNA.
Non-isotopic DNA probes:
For the production of non-isotopic DNA probes, one of the four deoxynucleotides (used for primer extension described above) is tagged with a label (e.g., biotin). The label of the DNA probes can be detected by use of chemical and enzymatic reactions.
2. Screening by Colony Hybridization:
The DNA sequence in the transformed colonies can be detected by hybridization with radioactive DNA probes (sometimes labeled RNA probes can also be used). Colony hybridization technique is also referred to as replica plating by some authors. The technique depicted in Fig. 9.7 is briefly described.
The transformed cells are grown as colonies on a master plate. Samples of each colony are transferred to a solid matrix such as nitrocellulose or nylon membrane. The transfer is carefully carried out to retain the pattern of the colonies on the master plate. Thus, the nitrocellulose paper contains a photocopy pattern of the master plate colonies. The colony cells are lysed and deproteinized.
The DNA is denatured and irreversibly bound to matrix. Now a radiolabeled DNA probe is added which hybridizes with the complementary target DNA. The non-hybridized probe molecules are washed away. The colony with hybridized probe can be identified on autoradiograph. The cells of this colony (from the master plate) can be isolated and cultured.
Many a times multiple colonies are detected on hybridization by a DNA probe. This is due to overlapping sequences. To identify which colony has the complete sequence of the target gene, data observed from the restriction endonuclease analysis will be helpful.
Modifications of Colony Hybridization Technique:
Several improvements in the colony hybridization technique, described above, have been made in recent years. In the plaque lift technique, nitrocellulose paper is directly applied on the upper surface of master agar plate making a direct contact. By this way, plaques can be lifted and several identical DNA prints can be made from a single plate. This technique increases reliability. More recently, screening of DNA libraries is carried out by automated techniques.
3. Screening by PCR:
Polymerase chain reaction (PCR) is as good as hybridization technique for screening DNA libraries. But adequate information (on the franking sequences of target DNA) must be available to prepare primers for this method. The colonies are maintained in multiwall plates, each well is screened by PCR and the positive wells are identified.
4. Screening by Immunological Assay:
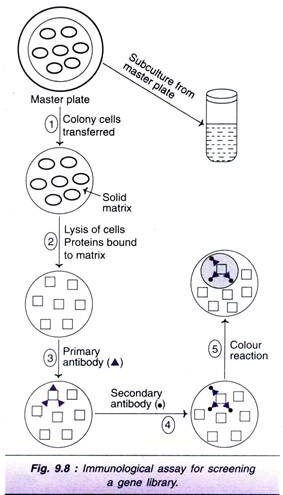
Immunological techniques can be used for the detection of a protein or a polypeptide, synthesized by a gene (through transcription followed by translation). The procedure adopted for immunological assay and hybridization technique (described already) are quite comparable. Screening procedure by immunological assay is depicted in Fig. 9.8, and briefly described hereunder.
The cells are grown as colonies on master plates which are transferred to a solid matrix (i.e., nitrocellulose). The colonies are then subjected to lysis and the released proteins bound to the matrix. These proteins are then treated with a primary antibody which specifically binds to the protein (acts as an antigen), encoded by the target DNA. After removing the unbound antibody by washings, a second antibody is added which specifically binds to the first antibody.
Again the unbound antibodies are removed by washings. The second antibody carries an enzyme label (e.g., horse reddish peroxidase or alkaline phosphatase) bound to it. The detection process is so devised that as a colourless substrate it is acted upon by this enzyme, a coloured product is formed. The colonies which give positive result (i.e., coloured spots) are identified. The cells of a specific colony can be sub-cultured from the master plate.
5. Screening by Protein Function:
If the target DNA of the gene library is capable of synthesizing a protein (particularly an enzyme) that is not normally produced by the host cell, the protein activity can be used for screening. A specific substrate is used, and its utilization by a colony of cells indicates the presence of an enzyme that .acts on the substrate. For instance, the genes coding for enzymes α-amylase and β-glucosidase can be identified by this technique.