Essay on Regulation of Gene Expression:- 1. Introduction to Regulation of Gene Expression 2. The Lac Operon 3. Bacteriophage Lambda (λ) 4. Gene Amplification during Development of Metazoans 5. Immunoglobulin Gene Rearrangement 6. Regulation of Messenger RNA Stability Provides another Control Mechanism.
Contents:
- Essay on Introduction to Regulation of Gene Expression
- Essay on the Lac Operon
- Essay on the Bacteriophage Lambda (λ)
- Essay on the Gene Amplification during Development of Metazoans
- Essay on the Immunoglobulin Gene Rearrangement
- Essay on the Regulation of Messenger RNA Stability Provides another Control Mechanism
1. Introduction to Regulation of Gene Expression:
A single ribosome is capable of translating about 400 codons in 10 seconds into a protein with a molecular weight of 40,000. Mammalian cells possess only about 1,000 times more genetic information than does the bacterium E. Coli. Much of this additional genetic information is probably involved in the regulation of gene expression.
There are only two types of gene regulation: positive regulation and negative regulation. When the expression of genetic information is quantitatively increased by the presence of a specific regulatory element, regulation is said to be positive’, whereas when the expression of genetic information is diminished by the presence of a specific regulatory element, regulation is said to be negative.
The element or molecule mediating the negative regulation is said to be a negative regulator, that mediating positive regulation is a positive regulator. A double negative has the effect of acting as a positive. Thus, an effector that inhibits the function of a negative regulator will appear to bring about a positive regulation.
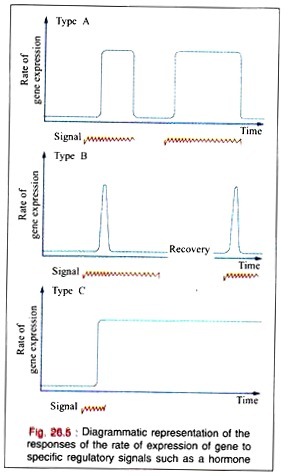
In many regulated systems that appear to be induced, they are depressed at the molecular level. In biologic organisms, there are three types of temporal responses to a regulatory signal. These three responses are shown in the Fig. 26.5 as rate of gene expression in temporal response to an inducing signal.
A Type A response is characterized by an increased rate of gene expression that is dependent upon the continued presence of the inducing signal. When the inducing signal is removed, the rate of gene expression is diminished to its basal level, but the rate repeatedly increases in response to the reappearance of the specific signal.
This type of response is commonly observed in many higher organisms after exposures to inducers such as steroid hormones.
A Type B response exhibits an increased rate of gene expression that is transient even in the continued presence of the regulatory signal. After the regulatory signal has terminated and the cell has been allowed to recover, a second transient response to a subsequent regulatory signal may be observed.
This type of response may commonly occur during development of an organism when only the transient appearance of a specific gene product is required although the signal persists.
The Type (response exhibits an increased rate of gene expression that persists indefinitely even after the termination of the signal. The signal acts as a trigger in this pattern. Once the gene expression is initiated in the cell, it cannot be terminated even in the daughter cells; it is, therefore, an irreversible and inherited alteration.
The gene ox cistron is the unit of genetic information. It is the smallest segment of the DNA molecule containing about 600 base pairs. The genetic message is carried in the sequence of bases along the DNA strand. These are arranged in orderly manner along the length of the DNA molecule in the chromosomes.
When a pair carries genes with the same characteristics, say tallness, then the individual is said to be homozygous with respect to that character. When one of the pair tallness and the other gene shortness, the individual is heterozygous.
2. The Lac Operon:
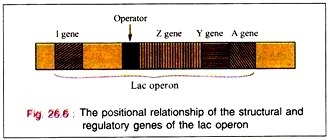
Lac operon is nothing but structural gene + operator gene.
A lactose analog which is capable of inducing the lac operon while not itself serving as a substrate for β-galactosidase is called a gratuitous inducer. The expression of some gene is constitutive i.e., they are expressed at a reasonably high rate in the absence of any specific regulatory signal.
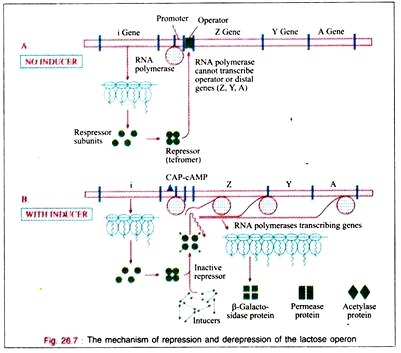
The regulator gene controls the activity of the operator gene. The regulator gene induces the synthesis of protein macromolecules (probably RNA protein) called repressors.
The operon becomes active because the repressor system is itself inactivated. The phenomenon is said to be De-repression. The operator locus is a region of double- stranded DNA of 27 base pairs long with a 2-fold rotational symmetry in a region that is 21 base pairs long.
The minimum effective size of an operator for lac repressor binding is 17 base pairs. The binding occurs mostly in the major groove without interrupting the base-paired double helical nature of the operator DNA.
The binding of the inductor to the repressor molecule involves the amino acid residues in positions 74 and 75. The operator locus lies between the Promoter site, at which the DMA- dependent RNA polymerase attaches to commence transcription, and the beginning of the Z gene.
When attached to the operator locus the repressor molecule prevents the transcription of the operator locus and of the distal structural genes, Z, Y, A. Thus, the repressor molecule is a negative regulator and in its presence the expression of the Z, Y, and A genes is prevented. Normally 20-40 repressor molecules are present and one or two operator loci per cell are also found to be present.
The translation of the polycistronic mRNA can occur even before the transcription is completed. The depression of the lac operon allows the cell to synthesize the enzymes necessary to catabolize lactose as an energy source. Only for the RNA polymerase to attach at the promoter site, there must be the presence of the catabolite gene activation protein (CAP) to which cAMP is attached.
The bacterium accumulates cAMP only when it is starved for a source of carbon. In the presence of glucose or glycerol in concentrations sufficient for growth, the bacteria will lack sufficient cAMP to bind to CAP.
Therefore, in the presence of glucose on glycerol, cAMP-saturated CAP is lacking, so that the DNA- dependent RNA polymerase cannot begin the transcription of the lac operon. In the presence of the CAP- cAMP complex on the promoter site transcription then takes place. Therefore, the CAP- cAMP regulator is acting as a positive regulator.
When the i gene is mutated so that its product, the lac repressor, is not capable of binding to operator DNA, the organism will exhibit constitutive expression of the lac operon.
An organism with an i gene mutation that prevents the binding of an inducer to the repressor will remain repressed even in the presence of the inducer molecule, because the inducer cannot bind to the repressor on the operator locus in order to depress the operon.
3. Bacteriophage Lambda (λ):
When a lamda infects a sensitive E. Coli, it injects its 45,000 base-pair, double stranded, linear DNA molecule into the cell (Fig. 26.8).
Depending upon the nutritional state of the cell, the lambda DNA will either integrate into the host genome (Lysogenic Pathway) and remain dormant until activated, or it will commence replicating until it has made about 100 copies of complete, protein-pack-aged virus at which point it effects lysis of its host (Lytic Pathway).
The newly generated virus articles can then infect other sensitive host.
When integrated into the host genome in its dormant state, lambda will remain in such a state until activated by exposure of its lysogenic bacterial host to DNA-damaging agents.
In response to such a noxious stimulus, the dormant bacteriophage becomes induced and begins to transcribe and subsequently translate those genes of its own genome which are necessary for its excision from the host chromosome, its DNA replication, and its protein cost and lysis enzymes. This event acts like a trigger of type C responses.
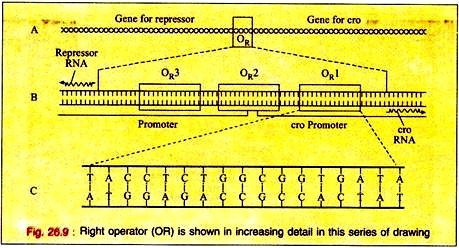
The switching event in lambda is centred around an eighty base pair region in its double- stranded DNA molecule referred to as the right operator (OR) Fig. 26.9A. The right operator is flanked on its left side by the structural gene for another regulatory protein called cro. When lambda is in its prophage state, the repressor gene is the only lambda gene that is expressed.
When the bacteriophage is undergoing lytic growth, the repressor gene is not expressed, but the cro gene, as well as many other genes in lambda, is expressed. That is,’ when the repressor gene is on, the cro gene is off, and when the cro gene is on, the repressor gene is off. These two genes regulate each other’s expression.
The operator region can be subdivided into three discrete sites, each consists of 17 base pairs of similar but not identical DNA sequence (Fig. 26.9B). Each of these 3 sub-regions, (OR1, OR2, and OR3) can bind either repressor or cro proteins in the major groove of the DNA double helix.
The DNA region between the cro and repressor genes also contains two promoter sequences that direct the binding of RNA polymerase in a specified orientation where it commences transcribing the adjacent genes.
One promoter directs RNA polymerase to begin transcription in the rightward direction and, thus, to transcribe cro and other distal genes, while the other promoter directs the transcription of the repressor gene in the leftward direction (Fig. 26.9B).
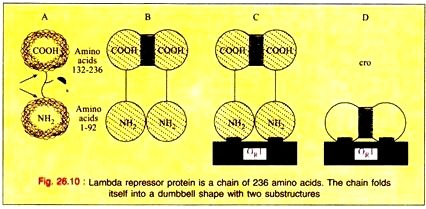
The product of the repressor gene, the 236 amino acid repressor protein, exists as a 2-domain molecule in which the amino-terminal domain binds to operator DNA and the carboxy-terminal domain promotes the association of one repressor protein with another to form a dimer.
A dimer of repressor molecules binds to operator DNA much more tightly than does the monomeric form (Fig. 26.10-A to C). The product of the cro gene, the 66-amino acid cro protein has a single domain but also binds the operator DNA more tightly as a dimer (Fig. 26.10D).
In a lysogenic bacterium i.e., a bacterium containing a lambda prophage, the lambda repressor dimer binds preferentially to OR1 but in doing so by a cooperative interaction, enhances the binding of another repressor dimer to OR2 (Fig 26.11). The affinity of repressor of OR1 is the least of the three operator sub-regions.
The binding of repressor to OR1 has two major effects. The occupation of ORI by repressor blocks the binding of RNA polymerase to the rightward Promoter and thereby prevents’ the expression of the cro gene. Secondly, repressor dimer bound to OR1 enhances the binding of repressor dimer OR2.
The binding of repressor to OR2 has the important effect of enhancing the binding of RNA polymerase to the leftward promoter that overlaps OR2 and thereby enhances the transcription and subsequent expression of the repressor gene.
Thus, the lambda repressor is both a negative regulator by preventing transcription of the cro gene, and a positive regulator, by enhancing the transcription of its own gene, the repressor gene. This dual effect of repressor is responsible for the stable state of the dormant lambda bacteriophage.
When a DNA damaging signal, such as ultraviolet light, strikes the lysogenic host bacterium, fragments of signal-stranded DNA are generated that activate a specific protease coded by a bacterial gene and referred to as rec A. The activated rec A protease hydrolyzes the portion of the repressor protein that connects the amino-terminal and carboxytenninal domains of that molecule.
Such cleavage of the repressor domains causes the repressor dimers to dissociate, which in turn causes a dissociation of the repressor molecules from OR2 and eventually from OR1. The effects of removal of repressor from OR1 and OR2 are speculated.
RNA polymerase immediately has access to the right- ward promoter and begins transcribing the cro gene, and the enhancement effect of the repressor at OR2 on leftward transcription is lost.
The cro protein translated from the newly transcribed cro gene also binds to the operator region as dimers, but its order of preference is the opposite of that of repressor. That is, cro binds most lightly to OR3, but there is no cooperative effect of cro at OR3 on the binding of Cro to OR2. At increasingly higher concentrations of Cro, the protein will bind to OR2 and eventually to OR1.
The occupancy of OR3 Cro immediately turns off the transcription from the leftward promoter and hence, prevents any further expression of the repressor gene. Therefore, the switch is completely effected. The Cro gene is now expressed and the repressor gene is fully turned off.
When Cro repressor concentration becomes very high, it will eventually occupy OR’ and in doing so turn down the expression of its own gene, a process that is necessary to effect the final stages of the lytic cycle.
4. Gene Amplification during Development of Metazoans:
During early development of metazoans, there is an abrupt increase in the need for specific genetic molecules such as ribosomal RNAs and messenger RNA molecules for proteins that make up such organs as the egg-shell. One way to increase the rate at which such molecules can be formed is to increase the number of genes available for transcription of these specific molecules.
Among the repetitive DNA sequences are thousands of copies of ribosomal RNA genes and tRNA genes. These genes preexist repetitively in the genomic material of the gametes and, thus, are transmitted in high copy number from generation to generation.
In some specific organisms such as the fruit fly (Drosophila), there occurs during oogenesis an amplification of a few pre-existing genes, such as these for the chorion (eggshell) proteins, S36 and S38. Subsequently, these amplified genes, presumably generated by a process of repeated initiations during DNA synthesis provide multiple sites for gene transcription.
In recent years, it has been possible to promote the amplification of specific genetic regions in cultured mammalian cells. In some cases, a several thousand-fold increase in the copy number of specific genes can be achieved over a period of time involving increasing doses of selective drugs.
In fact, it has been demonstrated in patients receiving methotrexate for treatment of cancer that malignant cells can develop drug resistance by increasing the number of genes for dihydrofolate reductase, the target of methotrexate.
5. Immunoglobulin Gene Rearrangement:
a. The coding segments responsible for the generation of specific protein molecules are frequently not contiguous in the mammalian genome. Immunoglobulin molecules consist of two types of polypeptide chains, light (L) and heavy (H) chains.
The L and H chains are each divided into N-terminal variable (V) and carboxy-terminal constant (C) regions. The V regions are responsible for the recognition of antigens (foreign molecules) and the constant regions for effector functions that determine how the antibody molecule will dispense with the antigen.
b. There are three unlinked families of genes responsible for immunoglobulin molecule structure. Two families are responsible for the chains (A, and k chains) and one family for heavy chains.
c. Each light chain is encoded by three distinct segments, the variable (VL), the joining (JL), and the constant (CL) elements. The mammalian haploid genome contains over 500 VL segments, five or six JL segments, and 10 or 20 CL segments.
d. A VL segment is brought from a distant site on the same chromosome to a position closer to the region of the genome containing the JL and CL segments during the differentiation of a lymphoid B cell. This DNA 4 rearrangement then allows the VL, JL and CL segments to be transcribed as a single mRNA precursor and subsequently processed to generate the mRNA for a specific antibody light chain.
The immunity system can generate a diverse library of antigen specific immunoglobulin molecules by rearrangement of the various VL, JL, and CL segments in the genome. The DNA rearrangement is referred to as V-J joining of the light chain.
e. The heavy chain is encoded by four gene segments the VH, the D (Diversity), the JH, and the CH DNA segment. The variable region of the heavy chain is generated by joining the VH with a D and a JH segment. The resulting VH-D-JH-DNA region is in turn linked to a CH gene. These CH genes (Cµ, Cδ, Cγ3 Cy1, Cy2b, Cy2α, Cα and Ce) determine the immunoglobulin class or subclass-lgM, lgG, IgA, etc. – of the immunoglobulin molecule.
f. A B cell that secretes antibody to a specific antigen will secrete antibodies of different classes having the same antigen specificity but different biologic roles during the differentiation. The different classes of immunoglobulin’s contain the same light chains and VH regions but different CH regions.
Thus, a single B cell and its derivatives can undergo class switching which is the result of second type of DNA rearrangement occurring during differentiation of the immunity system. The V-J joining for light chain expression and the V-D-J joining for heavy chain expression precede the class switching DNA rearrangement.
V-J Joining:
a. In the undifferentiated cell (e.g., germ line cell), the K-J gene (Jk) is closely linked to the Ck gene, but the gene segment for the k-variable region (Vk) is quite distant on the same chromosome.
b. Both the Vk-Jk and the similar Vλ-Jλ gene rearrangements to involve two short conserved sequence that exist in the direction 3′ to the V-segment and 5′ to the J- segment, close to the point of recombination.
c. There are two striking features of these conserved sequences. First, the conserved sequences of the JL segments are inverse complements of the conserved sequences in the VI. segments. Second, the length of the non-conserved sequence separating the heptamers and the monomers is highly conserved.
d. The variable region of the heavy chain involves three DNA segments, VH, D, and JH, which must be joined in a process involving two DNA rearrangements since all three segments are separated.
Class Switching:
1. Since differentiation proceeds and immunoglobulin production switches from IgM to IgA the V-D-J region of the parent B cell must be rearranged with C0 gene to permit the transcription of an mRNA precursor for an a-chain containing the same antigen-specific variable region.
2. The temporal order of the class switching’ is unidirectional within the physical order from left to right. Rearrangement of the CH genes seems to involve deletion of those CH genes 5′ to the CH gene joined to the V-D-J region.
The switch sites are different for different class switches but involve conserved sequences occurring in the appropriate regions of the genes to be rearranged.
Transcription Control:
a. Glucocorticoids regulate gene expression. Once they enter the mammalian cell, they bind to a steroid specific receptor molecule that undergoes a conformational change in the cytoplasm and enters the nucleus. The glucocorticoid receptor complex in the nucleus binds to a specific receptor-recognition site on DNA, a few hundred base pairs 5′ upstream from the transcription start site, for steroid-responsive genes.
The receptor recognition site on DNA influences the efficiency of utilization of the promoter by RNA polymerase and thereby influences the expression of the steroid responsive gene.
b. When an organism or its cultured cells are exposed to metal ions, such as zinc or cadmium, there is an accelerated rate of transcription of the metallothionein gene and a subsequent increase in the metallothionein protein to bind the potentially toxic heavy metal.
Another structural gene, such as that for thymidine kinase, can be ligated to this ‘metallothionein promoter region’ and the synthetic construct introduced to cultured cells, a small number of which will integrate the DNA into its own genome. When those cells are exposed to heavy metals, the metallothionein promoter region effects an induction of thymidine kinase.
c. There are many more primary transcripts in the nucleus than are represented as messenger RNA molecules in the cytoplasm. There must exist regulatory decisions as to which transcripts will ultimately be expressed and which will be discarded. There is no information available regarding the mechanism of this process.
6. Regulation of Messenger RNA Stability Provides another Control Mechanism:
a. Most mRN As in mammalian cells are very stable. Changes in the stability of a specific mRNA have the major effects in biologic processes.
b. Messenger RNAs exist in the cytoplasm as ribonucleoprotein particles (RNPs). Some of these proteins protect the mRNA from digestion by nucleases, while others under certain conditions, promote nuclease attack. Certain effectors, such as hormones, may regulate mRNA stability by increasing or decreasing the amount of the proteins.
c. Then ends of mRNA molecules are involved in mRNA stability. The 5′ cap structure in eukaryotic mRNA prevents attack by 5′ exonucleases, and the poly (A) tail prohibits the action of 3′ exonucleases. In mRNA molecules, a single endonucleolytic cut allows exonucleases to attack and digest the entire molecule.
Other structures in the 5′ noncoding sequence (5′ NCS) the coding region, and the 3’NCS are thought to promote or prevent this initial endonucleolytic action.
d. Structures at the 3′ end including the poly (A) tail enhance or diminish the stability of specific mRNAs. The absence of poly (A) tail is associated with rapid degradation of mRNA and the removal of poly (A) from some RNAs results in their destabilization.
e. In AU-rich regions, many of which contain the sequence AUUUA which appears in mRNAs that have a very short half-life including some encoding oncogene proteins and cytokines.