This article throws light upon the top three things to know about gene expression.
The top three things about gene expression are: (1) RNA Polymerase of Prokaryotes (2) Process of Transcription in Prokaryotes and (3) Process of Transcription in Eukaryotes.
Contents
Introduction:
Transcription is the process by which genetic information from DNA is transferred into RNA. DNA sequence is enzymatically copied by RNA polymerase to produce a complementary nucleotide RNA strand. Transcription produces different types of RNA molecules such as, mRNA, tRNA, rRNA and microRNA as described in chapter 5 tRNA and rRNA are involved in the synthesis of protein. RNA molecules are of different sizes depending upon the gene.
The amount of RNA produced from different genes also varies depending upon the requirement of the gene product as described in Chapter 9. RNA transcription in prokaryotes and eukaryotes is similar but several fundamental differences exist. The most striking difference is that prokaryotic cells lack a nucleus whereas it is present in eukaryotes. Therefore, in eukaryotes mRNA is transported from nucleus to the cytoplasm before the information within it can be converted into a protein product.
In the case of protein-encoding DNA, transcription is the first step that ultimately leads to the translation of the genetic code, via RNA intermediate, into a functional peptide or protein. The stretch of DNA that is transcribed into an RNA molecule is called a transcription unit. A transcription unit that is translated into protein contains sequence that directs and regulates protein synthesis in addition to coding sequence that is translated into protein.
Regulatory sequence that is before, or 5′, of the coding sequence is called 5′ un-translated (5’UTR) sequence, and sequence found following, or 3′, of the coding sequence is called 3′ un-translated (3’UTR) sequence. Transcription has some proofreading mechanisms, but they are fewer and less effective than the controls for copying DNA; therefore, transcription has a lower copying fidelity than DNA replication. Only one stand of DNA, called the template strand (antisense strand), acts as a template. The other strand is called as coding strand (sense strand).
As in DNA replication, transcription proceeds in the 5′ → 3′ direction. The DNA template strand is read 3′ → 5′ by RNA polymerase and the new RNA strand is synthesized in the 5′ → 3′ direction (Fig. 7.1). RNA polymerase binds to the 3′ end of a gene (promoter) on the DNA template strand and travels toward the 5′ end.
Except for the fact that thymine’s in DNA are represented as uracil’s in RNA, the newly synthesized RNA strand will have the same sequence as the coding (non-template) strand of the DNA. For this reason, scientists usually refer to the DNA coding strand that has the same sequence as the resulting RNA when referring to the directionality of genes on DNA, not the template strand. Transcription is divided into 3 stages: initiation, elongation and termination.
A molecule which allows the genetic material to be realized as a protein was first hypothesized by Jacob and Monod. RNA synthesis by RNA polymerase was established in vitro by several laboratories by 1965; however, the RNA synthesized by these enzymes had properties that suggested the existence of an additional factor needed to terminate transcription correctly. Recently, Roger D. Kornberg got the 2006 Nobel Prize in Chemistry “for his studies of the molecular basis of eukaryotic transcription”.
Thing # 1. RNA Polymerase of Prokaryotes:
RNA polymerase is the enzyme that is responsible for synthesis of RNA. It acts like DNA polymerase requiring a template, nucleotide tri phosphate (ATP, CTP, GTP, and UTP) and Mg2+ ions. The special characteristics of RNA polymerase is its ability to initiate chain growth without the need of a primer (DNA polymerase requires a primer to initiate DNA synthesis).
RNA polymerase from E. coli is composed of 6 polypeptide subunits-two alpha (α) subunits, one beta (β) subunit, one beta prime (β’) subunit, one omega (ω) subunit, one sigma (σ) subunit (Fig. 7.2). These five subunits are tightly bound together and constitute the core enzyme. The core enzyme in itself is capable of synthesizing RNA from DNA, but it is not capable of identifying the initiation site of a gene.
The sigma subunit is essential for initiating transcription at the beginning of a gene and binds to a core enzyme to form the holoenzyme. Core enzyme has 400kD molecular weight and four types of sigma units have 30-90 kD molecular weight.
Bacterial cells have several different sigma subunits and each is responsible for initiating RNA synthesis for a specific group of genes (Table 7.1). Each bacterial cell has ~4-7 thousands RNA polymerase molecules and 80-90% are engaged in active RNA synthesis in a rapidly dividing cells. A specific sigma subunit (in association with core enzyme) is able to recognize and binds to a specific DNA sequence called as promoter.
The promoter site defines the beginning of a gene for transcription. Consensus sequences located at -35 and -10 base pair upstream are recognized by sigma subunits. These sequences are quite similar in nucleotide arrangement.
In bacteria, promoter along with sigma subunit decides how frequently a gene is transcribed. For example, sigma serves to initiate transcription of housekeeping genes (those genes that are needed all the times to maintain the metabolism and structure of the cell) and thus initiates the synthesis of mRNAs from the largest class of genes.
However, there is variation in the promoter sequence among genes recognized by sigma subunit and this variation affects the frequency of transcription initiation. As a result, one gene may be transcribed with one initiation event every second, whereas, another may be transcribed every 5 minutes. This is a fundamental system of regulating the level of gene expression in prokaryotes.
Thing # 2. Process of Transcription in Prokaryotes:
Initiation:
Unlike DNA replication, transcription does not need a primer to start. RNA polymerase simply binds to the DNA and, along with other cofactors, unwinds the DNA to create an initiation bubble so that the RNA polymerase has access to the single-stranded DNA template.
Transcription initiation is far more complex in eukaryotes and archaea, the main difference being that eukaryotic polymerases do not recognize directly their core promoter sequences. Transcription factors must first mediate the binding of RNA polymerase and the initiation of transcription. The completed assembly of transcription factors and RNA polymerase bound to the promoter is called the transcription initiation complex.
All promoters recognized by a single sigma subunit have similar sequences, and nucleotides that occur most frequently in the promoter are known as a “consensus sequence”. The promoter consensus sequence is composed of two short sequences separated by about 20bp. These two groups of sequences are labeled the -35 and -10 sequences because they are situated 35 and 10 bases before the first base of DNA that will be transcribed into the first base of the RNA (Fig. 7.3).
DNA bases to the left (negative numbers) of the ‘start’ site are said to be ‘upstream’ and those on right side (positive numbers) are called as ‘downstream’. RNA polymerase binds to DNA in two steps. In first step, the enzyme binds to promoter. In 2nd step, the helix unwinds to allow base-pairs recognition to the template-the DNA sequence for the RNA polymerization reaction. DNA unwinding starts at the -10 sequence and proceeds to the right of the start point i.e., downstream. The sigma subunit is required only for recognition of the consensus sequence and thus detaches from the core enzyme.
The product of transcription is always RNA (tRNA, mRNA, rRNA, snRNA). The precursors for RNA synthesis are nucleotide triphosphates. The greatest varieties of RNAs are found in the mRNAs. To start RNA synthesis a ribonucleotide triphosphate is base paired at the RNA start site on the DNA. This represents the 5′ end of the new molecule and it grows when ribonucleotides are added to the 3′ end. The 5′ end retains the triphosphate precursor.
Elongation:
One strand of DNA, the template strand (or non-coding strand), is used as a template for RNA synthesis. As transcription proceeds, RNA polymerase traverses the template strand and uses base pairing complementarity with the DNA template to create an RNA copy. Although RNA polymerase traverses the template strand from 3′ 5′, the coding (non-template) strand is usually used as the reference point, so transcription is said to go from 5′ → 3′. This produces an RNA molecule from 5′ → 3′, an exact copy of the coding strand (except that thymines are replaced with uracils, and the nucleotides are composed of a ribose (5-carbon) sugar where DNA has deoxyribose (one less Oxygen atom) in its sugar-phosphate backbone).
In E.coli, the polymerization reaction occurs at a rate of 40 nucleotides per second at 37°C; thus a gene for 1000bp is transcribed in about 30 seconds. Finally, RNA polymerase will reach a terminator sequence that tells the enzyme to stop polymerization of the RNA and to dissociate from the DNA.
Unlike DNA replication, mRNA transcription can involve multiple RNA polymerases on a single DNA template and multiple rounds of replication, so many mRNA molecules can be produced from a single copy of a gene. This step also involves a proofreading mechanism that can replace incorrectly incorporated bases (Fig. 7.4).
 Termination:
Termination:
RNA synthesis is terminated by two mechanisms:
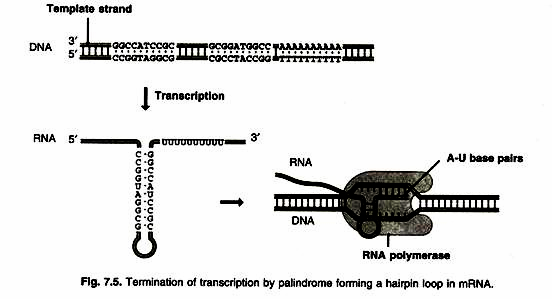
1. One type of termination sequence is composed of a stretch of AT nucleotides preceded by two symmetrical 8bp GC sequences (a palindrome) followed by short sequence of AT pairs (Fig. 7.5).
2. In other mechanism rho (p) protein terminates the synthesis.
In bacteria, a gene can be defined as physical entity-having a start site (promoter), the coding information and a terminator sequence. Another term for a gene is a cistron, which is the information required to make one polypeptide. Unfortunately, in bacteria organelles of eukaryotic cells, more than one polypeptide is encoded in a single message. These are called polycistronic mRNAs, because more than one cistron is present in mRNA. This is illustrated in detail in chapter 9 describing lac Operon and trp Operon.
Thing # 3. Transcription in Eukaryotes:
Three different RNA polymerases are found in all eukaryotic cells and each is responsible for the synthesis of a different class of RNA molecule. The most active RNA synthesizing abilities is associated with RNA polymerase I (RNA pol). Its product, ribosomal RNA, is a major component of ribosomes. The ribosome is a large ribonucleoprotein particle necessary for protein synthesis.
Many copies of rRNA must be made for protein synthesis to occur. Thus, RNA pol I accounts for most of the RNA synthesized in the cell. The second largest amount is produced by RNA pol III which synthesizes tRNA and 5s RNA, required for protein synthesis. RNA pol II is responsible for the synthesis of precursor of mRNA or heterogeneous nuclear RNA (pre RNA), which is usually much larger than mRNA. Pre RNA is enzymatically processed to form mRNA (Table 7.2).
Like prokaryotic genes eukaryotic genes have promoter and terminator sequences that direct the transcriptional machinery to start and stop. However, the signal sequences in the DNA for starting and stopping RNA synthesis are variable in eukaryotes. It is thus, impossible to specify a single sequence responsible for stop and start sequences for all genes.
Some common features are as follows:
1. In eukaryotes most promoters have a sequence called TATA box, which has the consensus sequence 5′-TATAAA-3′.
2. The TATA box is located about 25bp upstream (-25) from the starting point and determines the start site for transcription. It is surrounded by GC rich regions.
3. Two other sequences upstream of the start point also effect gene expression and are present in many eukaryotic promoters. These are called CAAT box and GC box. The CAAT box is named for its consensus sequence (GGCCAATCT) and located about 80bp upstream (-80).
4. Mutation in these sequences affects transcription.
RNA polymerase cannot bind itself and requires a group of proteins known as transcription factor. The eukaryotic transcription requires multiple protein binding sites of initiation of transcription and a number of transcription factors. In prokaryotes, RNA pol requires only sigma factor to bind to the core enzyme, which can then bind to promoter region (table 7.3).
In eukaryotes, promoters do not act alone in regulating the amount of RNA transcript synthesized. Another very important class of DNA sequences that control RNA transcription is the enhancer sequences or enhancer elements. Enhancer sequences are binding site for a wide variety of transcription factors.
The enhancer sequences in conjunction with the proper transcription factors can have an enormous effect on the level of synthesis of a RNA transcript, increasing it by as much as thousand-folds. Enhancer sequences are situated at highly variable distances from the gene that they control, sometimes several thousands base pairs from the promoter. These sequences can regulate the genes located upstream or downstream positions.
RNA Processing:
RNA processing is to generate a mature mRNA (for protein genes) or a functional tRNA or rRNA from the primary transcript. In this section, we discuss first the processing of pre-mRNA and then processing of pre-rRNA and pre-tRNA. In some cases, RNA editing is also involved.
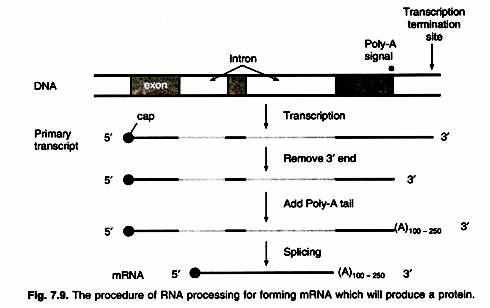
Processing of pre-mRNA involves the following steps:
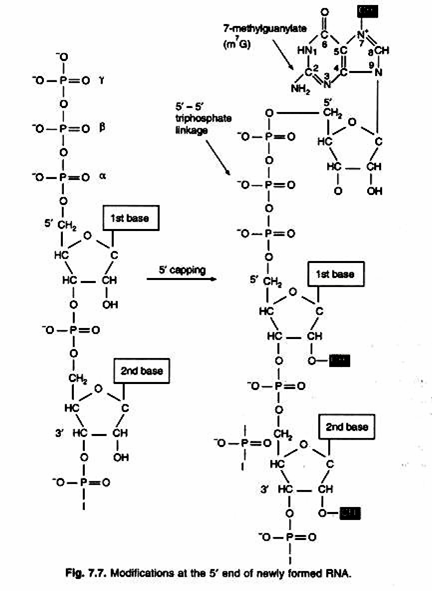
1. Capping – add 7-methylguanylate (m7G) to the 5′ end.
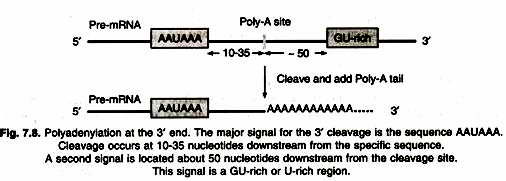
2. Polyadenylation – add a poly-A tail to the 3′ end.
3. Splicing – remove introns and join exons.
5′-Capping:
Capping occurs shortly after transcription begins. The chemical structure of the “cap” is shown in the following figure, where mG is linked to the first nucleotide by a special 5′-5′ triphosphate linkage. In most organisms, the first nucleotide is methylated at the 2′-hydroxyl of the ribose. In vertebrates, the second nucleotide is also methylated (Fig. 7.7).
3′-Polyadenylation:
A stretch of adenylate residues are added to the 3′ end. The poly-A tail contains ~ 250 A residues in mammals, and ~ 100 in yeasts (Fig. 7.8).
In general, introns tend to be much longer than exons. An average eukaryotic exon is only 140 nucleotides long, but one human intron stretches for 480,000 nucleotides. Removal of the introns – and splicing the exons together are among the essential steps in synthesizing mRNA (Fig. 7.9).
Splicing:
In genetics, splicing is a modification of genetic information after transcription, in which introns of precursor messenger RNA (pre-mRNA) are removed and exons of it are joined. Since in prokaryotic genomes introns do not exist, splicing naturally only occurs in eukaryotes. The splicing prepares the pre-mRNA to produce the mature messenger RNA (mRNA), which then undergoes translation as part of the protein synthesis to produce proteins. Splicing includes a series of biochemical reactions, which are catalyzed by the spliceosome, a complex of small nuclear ribonucleo-proteins (snRNPs).
Spliceosomal introns often reside in eukaryotic protein-coding genes. Within the intron, a 3′ splice site, 5′ splice site, and branch site are required for splicing. Splicing is catalyzed by the spliceosome which is a large RNA-protein complex composed of five small nuclear ribonucleoproteins (snRNPs, pronounced ‘snurps’). The RNA components of snRNPs interact with the intron and may be involved in catalysis. Two types of spliceosomes have been identified (the major and minor) which contain different snRNPs.
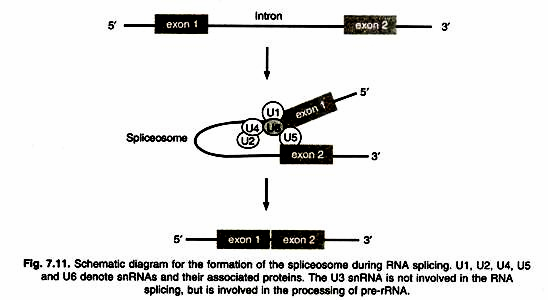
It involves five snRNAs and their associated proteins. These ribonucleoproteins form a large (60S) complex, called spliceosome. Then, after a two-step enzymatic reaction, the intron is removed and two neighboring exons are joined together.
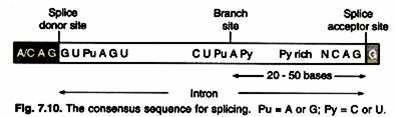
The branch point A residue plays a critical role in the enzymatic reaction. In removing introns from pre mRNA, the enzyme responsible for the breakage and reformation of nucleotide- nucleotide bonds recognize conserved (consensus) sequence present at intron-exon junctions (Fig. 7.10). The major spliceosome splices introns containing GU at the 5′ splice site and AG at the 3′ splice site. It is composed of the U1, U2, U4, U5, and U6 snRNPs (These named U1, U2 etc because of abundance of uridines).
The GU-AG rule is nearly always obeyed. The presence of a conserved sequence implies a common mechanism for splicing in eukaryotes. The first step of splicing reaction involves the enzymatic cleavage of the left junction site (just before GU in the RNA sequence) followed by formation of a lariat structure between the release 5′ end of the intron and the 2′ carbon in the sugar ring of an adenosine within the intron (Fig. 7.11).
The right junction is then cleaved, releasing the branched lariat and simultaneously joining the two exons. The released intron RNA fragment is degraded and the nucleotides are recycled into new RNA. The splicing reaction is catalyzed by a large complex called spliceosome, which consists of nucleic acids and 8-10 enzymes.
Self-Splicing or Autocatalytic Splicing:
Self-splicing occurs for rare introns that form a ribozyme, performing the functions of the spliceosome by RNA alone. There are two kinds of self-splicing introns, Group I and Group II. Group I and II introns perform splicing similar to the spliceosome without requiring any protein. This similarity suggests that Group I and II introns may be evolutionarily related to the spliceosome.
Self-splicing may also be very ancient, and may have existed in an RNA world that was present before protein. The splicing mechanism requires 5 additional RNA molecules and over 50 proteins are used and hydrolyzes many ATP molecules. The splicing mechanisms use ATP in order to accurately splice mRNAs. If the cell were to not use any ATPs, the process would be highly inaccurate and many mistakes would occur.
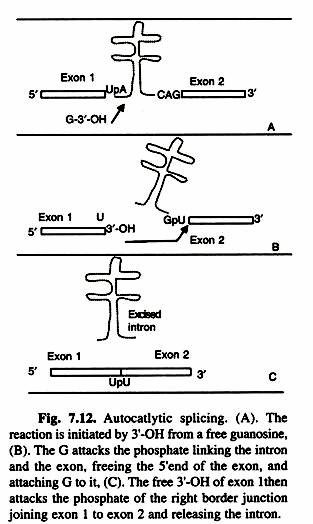
Two transesterfications characterize the mechanism in which group I introns are sliced: 1) 3’OH of a free guanine nucleoside (or one located in the intron) or a nucleotide cofactor (GMP, GDP, and GTP) attacks phosphate at the 5′ intron splice site. These consensus sequences can be located at a distance from intron-exon junctions.
Double starnded regions form between intron consensus sequences. The folded secondary structure allows an attacking guanosine residue to release side of intron, joining and guiding the two intron-exon junctions in close proximity. The guanosine becomes linked to the intron, and the free 3′-hydroxyl on the left exon then attaches to the right 3′ exon- intron junction and releasing the intron (Fig. 7.12).
3’OH of the 5′ end (exon 1) and 5’OH of exon 1 2 becomes a nucleophile and the second transesterfication results in the joining of the two exons. Autocatalytic introns splicing is found in the nuclear genes coding for rRNA in Tetrahymena and Physarum (a ciliate and a slime mold), in the mitochondria of fungi, and in mitochondria and chloroplasts of many plants. There are RNAs capable of self splicing. Catalysis by RNA may be quite wide spread and may have significant evolutionary implications. Such RNA molecules are called ribozymes.
A number of ribozymes are able to cleave a phosphodiester bond Rhibozyme linking nucleotide in an RNA strand. RNA-cleaving ribozymes provide on strategy to disrupt the function of specific mRNA. These ribozyme cleave at specific sequences and make double stranded structures by base pair complementation. Such strategy can be used to destroy RNA produced by a pathogen, e.g., AIDS virus (Fig. 7.13).
Evolution:
Splicing occurs in all the kingdoms or domains of life, however, the extent and types of splicing can be very different between the major divisions. Eukaryotes splice many protein-coding messenger RNAs and some non-coding RNAs. Prokaryotes, on the other hand, splice rarely, but mostly non-coding RNAs. Another important difference between these two groups of organisms is that prokaryotes completely lack the spliceosomal pathway.
Protein Splicing (Inteins):
Not only pre-mRNA but also proteins can undergo splicing. Although the bio-molecular mechanisms are different, the principle is the same, that parts of the protein, called inteins instead of introns, are removed. The remaining parts, called exteins instead of exons, are fused together. However, protein splicing has so far not been observed in humans, but in yeast.
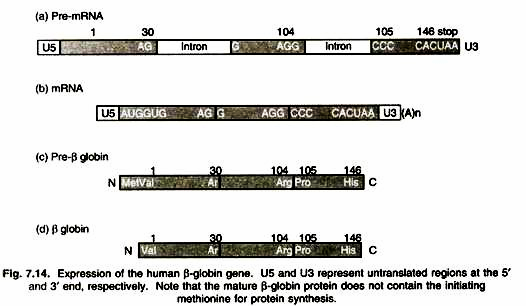
β-globin Gene:
Expression of the (3-globin gene is a typical process. This gene contains two introns and three exons. Interestingly, the codon of the 30th amino acid, AGG, is separated by an intron. As a result, the first two nucleotides AG are in one exon and the third nucleotide G is in another exon (Fig. 7.14).