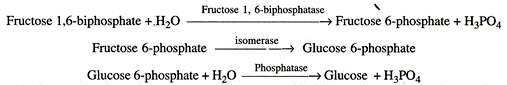
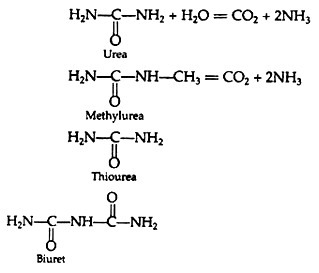
Let us make an in-depth study of informational macromolecules.
Macromolecule of the cell can, conveniently be defined as, polymers of high molecular weight, assembled from relatively simple precursors. Many of the molecules found within cells, like the polysaccharides, proteins and nucleic acids are macromolecules. Macromolecules have molecular weights from tens of thousands to billions.
Proteins have molecular weights ranging from 5000 to over 1 million; the nucleic acids have molecular weights up to several billions; and polysaccharides have molecular weights up to the millions. Macromolecules are produced by the polymerization of relatively small subunits with molecular weights of 500 or less.
Macromolecules themselves may be further assembled into supra-molecular complexes, membranes and organelles. Out of the three macromolecules, the polysaccharides i.e. polymers made up of a single kind of unit or two different alternating sugar units, serve as energy-yielding fuel stores and as extracellular structural elements, hence are not informational macromolecules. On the other hand the major tools of molecular biology and biotechnology are the proteins and nucleic acids and are known as the informational macromolecules.
Gene:
A chromosomal (DNA or RNA in some) segment that codes for a single polypeptide chain or RNA molecule is known as a gene.
One amino acid in a polypeptide chain is coded by a sequence of three consecutive nucleotides of a single strand of DNA (or RNA) and this sequence of three nucleotides (in mRNA) is called the genetic code. Thus the number of nucleotides in a gene for a particular polypeptide will be three times the number of the amino acids present in it.
In prokaryotes there are two types of genes:
(a) Structural genes:
Are those which specify the final gene product i.e. a protein or RNA.
(b) Regulatory genes:
Are those genes which control the structural genes. All the proteins/RNA are not always required by the cell. Some of the proteins/RNA are required at one time and yet some others at other times. At a given time some are required in lesser quantities and at other times in larger quantities.
In order to control the level of these proteins/RNA in the cell, the functioning of structural genes is controlled by the regulatory genes, which composed of three regions called (i) operator site-O, (ii) promoter site-P and (iii) repressor/inhibitor/initiator site-I. Further, a gene, in biological sense is a portion of a chromosome that determines or affects a single character or phenotype.
Genome:
All the genes and inter-genic DNA of an organism or an individual are collectively known as ‘genome’. The genome constitutes the complete chromosomes of an organism. Many virus and bacterial cells have a single chromosome and eukaryotes have many chromosomes. The number of genes in a viral genome like φ174 is 9 genes.
The E. Coli genome (chromosome) is about 4,400 genes and the human genome consists of about 1, 00,000 different genes. There is a difference between the viral (particulate) and cellular genome. In virus the genome has genes within genes or overlapping genes i.e. part of one gene will also be a part of another gene.
Likewise many genes in viral genome share the bases. φX174 genome has nine genes (A to J). Gene B is within the sequence of gene A and Gene E is within gene D.
In eukaryotes many genes contain intervening non-translated sequences called introns. The translated sequences are called exons. Due to these alternating introns and exons, the co-linear relationship between the nucleotide sequence of the gene and the amino acid sequence of polypeptide it codes, is lost.
Introns vary in number, position and fraction of the total length of the gene they occupy. Generally, the introns of a gene are much longer (60-80%) than the exons (20-40%). There are 50 introns in the collagen gene. Likewise ovalbumin has 7 introns and cytochrome-b gene has 4 introns.
Palindromes:
A segment of duplex DNA in which the base sequence of the two strands exhibit two fold rotational symmetry about an axis.
This is just like a sentence which is spelled the same way, when read forward or backward.
For example-
The palindromic sequences are self-complementary within each of the strands. Due to this complementarity, intra-chain base pairing is possible giving rise to a hairpin or cruciform structure.
The same type of hairpin is possible in the other strand also and at the same point of rotational symmetry. This results in a cruciform structure. Many such palindromes occur in each genome. The hairpin and cruciform structures help in functioning of DNA like termination of transcription.
Restriction modification/restriction endonucleases:
Most of the bacteria have a special type of mechanism which protects its own DNA, whereas destroys foreign DNA i.e. the DNA of invading viruses. This is brought about by a system of complex enzymes called restriction modification system.
This system contains two types of enzymes:
(a) DNA methylase and
(b) Restriction endonucleases.
Some of the bases in the genome are methylated by the enzyme DNA methylase and thus modifies the nucleotides at specific sequences in a particular bacterium. These sequences are short and palindromic. The restriction endonucleases recognise these sequences and cleave at that very point of those DNA which are not methylated. Thus the cell’s own DNA is not cleaved, whereas it recognises and cleaves the foreign DNA. This cleaved DNA is called “restricted DNA”.
More than 800 restriction endonucleases are discovered so far, in different bacterial species. Each enzyme (in some cases more) can recognise 100 different specific sequences. There are three types of restriction endonucleases designated as type—I, II & III. The name of each enzyme consists of a three letter abbreviation of the bacterial species from which it is derived. The following are the recognition sequences for some type—II restriction endonucleases showing methylated bases (*) and the site of cleavage (↑).
Note that by the action of EcoRI and Hindlll, the resulting DNA will be in two pieces, such that there are 2 to 4 nucleotides of one strand unpaired at each resulting end. These are referred to as cohesive ends or sticky ends, because they can base pair with each other or with complementary sticky ends of other DNA fragments.
On the other hand the action of PvuII and Haelll, cleaves both strands of DNA resulting in no unpaired bases. These are called blunt ends.
Restriction endonucleases are the key tools of recombinant DNA technology and thereby of Biotechnology.
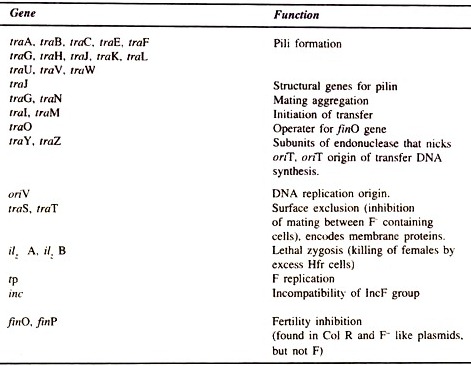
Plasmids:
Plasmids are extra chromosomal, independently replicating small circular DNA molecules. Plasmids are found in yeast, other fungi and in bacteria.
Naturally occurring bacterial plasmids range in size from 5000 to 400000 base pairs. The number of plasmid in each cell may be one or more than one (10 to 50 copies/cell). Generally, the plasmids remain
separately and detached from the chromosomal DNA. Plasmids carry genetic information and undergo replication to yield daughter plasmids, which pass into the daughter cells at cell division.
Plasmids do not have any role in the physiological functions of the organism in which they exist. Plasmids carry the gene for making the bacterium resistant to antibacterial agents (antibiotics), ex. plasmids carry gene for the enzyme beta-lactamase and thus makes that bacterium resistant to beta lactum antibiotics like amoxicillin and penicillin.
These plasmids can also pass from an antibiotic resistant cell to an antibiotic sensitive cell of the same or another bacterial species, thus making the other cell also resistant. Plasmids are very useful tools of modern biotechnology.