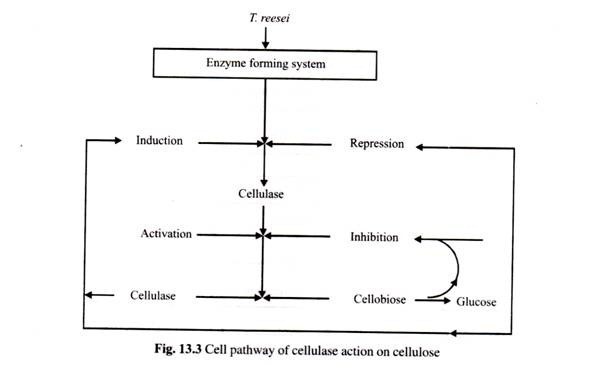
This essay deals with the restriction enzymes and other useful enzymes which are commonly used in genetic engineering.
Enzymes Used in Genetic Engineering
Essay Contents:
- Essay on the Discovery of Enzymes
- Essay on Restriction Endonuclease Enzyme
- Essay on Ligase Enzymes
- Essay on Alkaline Phosphatase Enzyme
- Essay on Phosphonucleotide Kinase Enzyme
- Essay on S1 Nuclease Enzyme
- Essay on DNA Polymerase I: Holoenzyme
- Essay on DNA Polymerase I: Klenow Fragment
- Essay on T4 DNA Polymerase Enzyme
- Essay on Taq DNA Polymerase Enzyme
- Essay on Ribonuclease
- Essay on Deoxyribonuclease I (Dnase I)
- Essay on Terminal Deoxynucleotidyl Transferase Enzyme
- Essay on Reverse Transcriptase Enzyme
Essay # 1. Discovery of Enzymes:
Genetic engineering was born because scientists learned to manipulate DNA. This skill was derived mainly from the field of nucleic acid enzymology. Prior to 1970, there was simply no technique available for cutting a duplex (double-stranded) DNA molecule into distinct fragments. Discovery of DNA metabolising enzymes granted scientists to propose and initiate genetic engineering.
All sort of DNA research developed from the ability, to cut DNA molecules at defined sequences. In other words, it was based upon the discovery of type II restriction endonuclease enzymes.
The isolation of the first restriction endonuclease enzymes, such as Hind II and Hind III, was a result of an interesting discovery by Hamilton O. Smith and his coworkers (1971) that Haemophilus influenza extracts contained activities that cut large DNA molecules into defined fragments.
H. influenza is a non-motile, gram-negative, facultative, anaerobic, pathogenic, rod shaped bacterium which is associated with human respiratory infections, conjunctivitis and meningitis.
Smith’s discovery gave rise to recombinant DNA research. In addition, an entire industry developed with the main purpose of discovery, characterization, purification and marketing of over 100 different site-specific restriction enzymes.
The bringing together of DNA fragments to form covalently linked chimeric molecules is the basis of recombinant DNA research. This step is essential in genetic engineering. This is attained by ligation which is catalysed by DNA ligase, an enzyme which was discovered much prior to that of restriction enzymes.
Before 1970 the existing central dogma in molecular biology was that genetic information transfer occurred from DNA to RNA, and then to protein. The proof that RNA-to-DNA information transfer did occur is based on the discovery and characterization of reverse transcriptase enzyme by Temin and Baltimore (1970).
Reverse transcriptase enzyme allows scientists to generate DNA copies (cDNA) of mRNA subsequent to cloning. The generation of cDNA, containing direct protein coding information is the normal step in cloning of eukaryotic genes.
In fact the most revolutionary and most simplistic molecular biological technical development, is PCR (i.e., polymerase chain reaction). PGR is a direct application of DNA polymerase to permit the test tube binomial amplification of specific DNA sequences.
The discovery of type II restriction enzymes demonstrated the enormous power and utility of site specific DNA cleavage reagents. This article deals with the restriction enzymes and other useful enzymes which are commonly used in genetic engineering (Table 55.1).
Essay # 2. Restriction Endonuclease Enzyme:
Restriction endonucleases (RE) are special class of endonucleases which cleave DNA molecules only at specific nucleotide sequences, called restriction sites. These specific sequences are of four to six nucleotides.
A tetranucleotide sequence will occur more frequently in a given molecule than hexanucleotide, therefore, more fragments will be produced by an enzyme which recognizes tetranucleotide sequence. At these restriction sites, restriction endonucleases cut the DNA by cleaving two phosphodiester bonds one within each strand of the double stranded DNA.
The term ‘restriction endonuclease’ was coined by Lederberg and Meselson (1964) to describe the nuclease enzymes that destroy (‘restrict’) any foreign DNA entering the host cell. However, the first restriction endonuclease enzyme to be isolated and studied was E. coli K12 by Meselson and Yuan (1968). Now these enzymes have been classified into three different types viz., Type I, Type II and Type III.
The discovery of these enzymes led to Nobel Prize for W. Arber, H. Smith and D. Nathans in 1978. In gene manipulation technology, restriction endonuclease enzymes are popularly called molecular knives, molecular scissors or molecular scalpels.
Essay # 3. Ligase Enzymes:
While exact cutting of DNA molecule is very useful for DNA cloning, its full potential is only exhibited when the fragments produced are joined together to give a new structure, known as recombinant DNA. This joining or ligation is achieved by the use of a DNA ligase enzyme.
The most common ligase enzyme is isolated from the bacterial virus (i.e., T4 bacteriophage). Thus, DNA ligases form a group of enzymes which mediate annealing, sealing or joining of DNA fragments. Primarily, ligase enzymes are involved in the repair of DNA molecule where sealing or union of DNA fragments takes place.
DNA ligases also play active part in processes such as DNA replication and recombination. These enzymes are widely used in genetic engineering for the production of hybrid DNA. Since ligase enzymes join DNA fragments or seal the nicks in the chain, they are called molecular structures.
Activity of DNA Ligase Enzymes:
(i) Ligation of DNA molecules with sticky or cohesive ends:
If two different DNA preparations are treated with the same restriction enzyme to give fragments with sticky ends, these ends will be identical in both preparations. Thus, when the two sets of DNA fragments are mixed, base pairing between sticky ends will result in the coming together of fragments which were derived from different molecules.
Also there will be pairing of fragments derived from the same molecule. Such pairing are temporary, owing to the weakness of hydrogen bonding between the few bases in the sticky ends.
The pairing can be stabilised by the use of DNA ligase, which forms a covalent bond between the 5′-phosphoryl at the end of one strand and 3′-hydroxyl of the adjacent strand. Polynucleotide ligase enzyme of T4 bacteriophage catalyzes the end to end joining of DNA duplexes at the base paired end. This reaction could occur intermolecularly or intramolecularly. Researches confirm that intermolecular mode of reaction is correct.
The ligation reaction is driven by ATP and is carried out at 4°C to lower the kinetic energy of molecules. This reduces separation of paired sticky ends and are later stabilised by ligation. However, long reaction time is required to compensate for the low activity of DNA ligase enzyme in the cold. The enzyme concentration is kept high and polyethylene glycol is added to reaction mixture for Stimulation.
Since ligation reconstructs the site of cleavage, recombinant DNA molecules produced by ligation of sticky ends can be cleaved again at the joints, using the same restriction endonuclease enzymes that was used to generate the fragment initially. As a result, a fragment can be inserted into a vector DNA, and recovered again after cloning of the recombinant molecules.
(ii) Ligation of DNA molecules with blunt ends:
Fragments of blunt-ended DNA can be ligated, but since there is no base-pairing to hold fragments together temporarily, concentrations of DNA and ligase enzyme must be high. However, blunt-end ligation is a useful way of joining together DNA fragments which have not been produced by the same restriction enzyme, and which therefore have mismatched sticky ends. These ends are removed prior to ligation, using the enzyme S1 nuclease, which digests single-stranded DNA.
In case of ligation of blunt-ends, a restriction site will not be regenerated and this may prevent recovery of a fragment after cloning. For this reason, short DNA duplexes, called linkers, are frequently used for joining DNA.
Linkers are short, double-stranded oligonucleotides, with blunt ends, containing at least one restriction site (i.e., Eco RI palindrome) within their sequence. These linkers can be joined to one preparation of DNA by blunt- ended ligation and then sticky ends can be created by cleavage of the linkers with a suitable restriction enzyme.
The linker is chosen so that the sticky end it produces is identical to that on the other DNA preparation. Consequently, the two can then be joined by ligation of their sticky ends. Some very versatile linkers are available which contain restriction sites for several different enzymes within a sequence of only eight to ten nucleotides. Blunt- ended molecules can also be ligated after building sticky ends.
Thus, with this method it is now possible to insert a foreign DNA segment at a particular site in the linker region of the vector and then retrieve this foreign DNA segment whenever necessary.
Sources of DNA Ligases:
DNA ligases are isolated from E. coli and T4 bacteriophage. The ligase enzyme isolated from E. coli is a polypeptide chain with a molecular weight of 75 kDa. It requires NAD+ as cofactor. The ligase obtained from T4 bacteriophage is 68 kDa. It requires ATP as a cofactor and a source of energy.
For the routine laboratory requirement, T4 DNA ligase is obtained from an induced lysogen of lambda T4 lig phage. This enzyme has the capacity to ligate a variety of cohesive and blunt- ended DNA fragments. The enzyme concentration is kept higher and a fusogen called polyethylene glycol, is added to reaction mixture for stimulation.
Application of DNA Ligase Enzymes:
DNA ligase enzymes play an important role in genetic engineering. In the absence of DNA ligase enzymes, recombinant DNA technology cannot be successful.
The important functions of DNA ligase enzymes are as follows:
1. Genetic engineering experiments involve joining of DNA fragments to produce recombinant DNA molecules. Ligase enzymes are used in the joining process.
2. Ligase enzymes help in ligation of vector and inserting recombinant DNA.
3. They help in ligation of linkers or adapter molecules at the blunt ends of DNA fragments.
4. They help in sealing nicks in double- stranded DNA.
5. Ligase enzymes requires 3′ OH and 5′ P0^4 group for ligation.
This requirement can be advantageous as:
(i) Self ligation of DNA can be prevented by dephosphorylation (using alkaline phosphatase) of intended donor fragments prior to ligation.
(ii) Dephosphorylation of vector DNA will prevent recircularization of the vector in cloning procedure.
Essay # 4. Alkaline Phosphatase Enzyme:
The enzyme alkaline phosphatase (AP) catalyses the removal of the 5′-terminal phosphate residues from nucleic acids (RNA, DNA and ribo- and deoxyribonucleotide triphosphates). This enzyme is isolated from bacteria (BAP) or calf intestine (CAP).
This enzyme is a dimeric glycoprotein with a molecular weight 14,000. It is made up of two identical or similar subunits each with a molecular weight of 6900. It is a zinc-containing enzyme with four atoms of Zn2+ per molecule.
Uses of Alkaline Phosphatase Enzyme:
1. Linearized cloning vectors can be prevented from recircularizing by dephosphorylation with alkaline phosphatase enzyme.
2. The free 5′-OH can be phosphorylated with polynucleotide kinase and ϒ-32P ATP to produce 32P end labelled nucleic acid.
3. AP enzyme is used for mapping and DNA fingerprinting studies.
Essay # 5. Phosphonucleotide Kinase Enzyme:
The enzyme phosphonucleotide kinase catalyses the transfer of the terminal phosphate group of ATP to the 5′-hydroxylated terminal of DNA or RNA. This enzyme is frequently used to end- label the nucleic acids with 32P (i.e., it adds the phosphate back to 5′-termini of DNA).
This can be accomplished by any method among following:
1. Forward reaction:
Transfer of labelled ϒ-phosphate form (ϒ-32P)-ATP to the free 5′-hydroxyl group of substrate-
5′ – OH – DNA + [-32P] ATP5′32 → PO – DNA + ADP
Substrate lacking a free 5′-hydroxyl requires prior dephosphorylation by alkaline phosphatase.
2. Exchange reaction:
In the initial step, the terminal 5′-phosphate is transferred from substrate to ADP present in the reaction mixture. Then, the labelled ϒ-phosphate from [ϒ-32P]-ATP is transferred to free hydroxyl group of substrate.
5′ – PO – DNA + ADP → 5’HO – DNA + ATP
5′ – HO – DNA + [ϒ-32 P] – ATP → 5′32 PO – DNA + ATP
Uses of Polynucleotide Kinase Enzyme:
The enzyme polynucleotide kinase is used to label 5′-termini of DNA and RNA with [ϒ-32P]-ATP by phosphorylation of 5′-hydroxyl groups or by the exchange reaction. This 5′-terminal labelling is used in mapping of restriction sites, DNA or RNA fingerprinting, hybridization studies and sequence analysis of DNA.
Essay # 6. S1 Nuclease Enzyme:
The S1 nuclease enzyme is single- strand specific endonuclease which cleaves DNA to release 5′-mono and 5′-oligonucleotides. Normally, double- stranded DNA, double- stranded RNA and DNA-RNA hybrids are resistant to action of S1 nuclease enzyme.
However, very large amounts of S1 nuclease enzyme can completely hydrolyze double- stranded nucleic acids. The enzyme hydrolyzes single stranded regions in duplex DNA such as loops and gaps.
S1 nuclease enzyme can also cleave single stranded areas of super helical DNA at torsional stress points where DNA may be unpaired or weakly hydrogen bonded. Once the super-helical DNA is nicked, S1 nuclease enzyme can cleave the second strand near the nick to generate linear DNA.
S1 nuclease enzyme is a monomeric protein with 3800 dalton molecular weight. It requires Zn2+ for its activity and is relatively stable against denaturing reagents such as urea, SDS and formamide. The optimum pH requirement lies between 4 to 4.5.
Uses of S1 Nuclease Enzyme:
1. S1 nuclease enzyme is used to analyse DNA-RNA hybrid structures to map transcripts.
2. It can be used to remove singles stranded tails from DNA fragments to produce blunt ends.
3. Hair pin loop structures formed during synthesis of double-stranded cDNA is digested by this enzyme.
4. S1 nuclease enzyme is also used for DNA mapping, called SI nuclease mapping Turner.
Essay # 7. DNA Polymerase I: Holoenzyme:
This enzyme has two-fold activities:
5′ → 3’exonuclease activity and DNA synthesis (acts as 5’ -3’ polymerase). Such a bifunctional activity enables the DNA polymerase I enzyme to use nicks or gaps in double- a stranded DNA as a starting point of DNA synthesis.
The 5′-exonuclease activity degrades that DNA strand which is complementary to the template strand and thus forming nick. DNA synthesis begins at 3′-end of nick and produces a new strand of DNA complementary to the template.
The new result is the movement of nick along the template strand (nick translation) until all the DNA complementary to the template strand (starting from the site of the origin of nick to the 5′-end of the template strand) is replaced.
Uses of DNA Polymerase I:
1. DNA polymerase I enzyme is used with radioactive or biotinylated nucleotides to prepare labelled DNA of high specific activity.
2. DNA polymerase I enzyme can also catalyse de novo DNA synthesis.
3. The enzyme DNA polymerase I has 3’→5′ proof-reading exonuclease activities on a single polynucleotide chain.
Essay # 8. DNA Polymerase I: Klenow Fragment:
Treatment of DNA polymerase I holoenzyme of E. coli with protease enzyme results in the production of two protein fragments. The larger fragment is called Klenow fragment and it does not show 5′-exonuclease activity (i.e., the 5′- exonuclease activity is exhibited by the intact enzyme) This Klenow fragment is used to synthesize DNA when there is no need of removing the DNA strand which is complementary to the template strand.
Uses of Klenow Fragment:
Klenow fragments are used in the following ways:
1. In DNA sequencing by dideoxy method.
2. For the production of second strand of cDNA.
3. Radiolabelling by filling in 5′-single stranded extension on double-stranded DNA.
4. Mutagenesis of DNA with synthetic oligonucleotides.
5. In labelling the DNA by random primer method.
+ stands for presence of particular activity by the enzyme.
0 presents absence of such activity.
Essay # 9. T4 DNA Polymerase Enzyme:
The enzyme T4 DNA polymerase lacks 5′-exonuclease activity but has a very active 3′- exonuclease action. This property allows radiolabelling of DNA fragment by replacement synthesis.
In method of replacement synthesis, T4 DNA polymerase enzyme acts on both the 3’ -ends of double sanded extensions. DNA complementary to these single strands are synthesized with deoxynucleoside triphosphate enzyme in the presence of radiolabelling compounds.
Application of T4 DNA polymerase:
1. T4 DNA polymerase is useful in generating 5′ single-stranded ends.
2. The enzyme can be used in radiolabelling of DNA.
Essay # 10. Taq DNA Polymerase Enzyme:
The enzyme Taq to DNA polymerase is isolated from the thermophilic bacterium Thermus aquaticus. Taq enzyme has the highest DNA polymerase activity at a pH of 9 and temperature around 75°C.
A Activity of Taq DNA polymerase is resistant to incubation at high as 95°C. Taq enzyme consists of a single polypeptide chain with a molecular weight of 95000. It lacks 5′ to 3′ and 3′ to 5′ exonuclease activity.
The highly thermostable Taq DNA polymerase from Thermus aquaticus is ideal for both manual and automated DNA sequencing because it is fast, highly progressive, has little or no 3’ – exonuclease activity and is active over a broad range of temperature.
Application of Taq Polymerase:
1. Taq enzyme is used in DNA sequencing studies.
2. Taq enzyme is used in ‘Polymerase chain reaction’ or PGR as it can withstand high temperatures.
Essay # 11. Ribonuclease:
Generally RNase A and RNase T1 enzymes are used in genetic engineering techniques. Both enzymes cleave the phosphodiester bond between adjacent ribonucleotides. RNase A cleaves next to uracil (U) and cytosine (C) in such a way that phosphate remains with these pyrimidines. The nucleotide present on the other side of phosphate is dephosphorylated. RNase A enzyme is isolated from the bovine pancreas.
RNase T1 cleaves specifically next to guanine. The phosphate group at the 3′ end of the nucleotide remains with the cut end. This enzyme is isolated from Aspergillus oryzae.
Ribonuclease H (RNase H):
The enzyme RNase H is an endoribonuclease that degrades the RNA portion of the RNA- DNA hybrids. RNase H enzyme cuts the RNA into short fragments.
Applications of RNase H:
1. RNase H is the key enzyme in the cDNA cloning technique. In this case, it is used to remove the mRNA from the RNA-DNA hybrid.
2. RNase H enzyme is used to detect the presence of RNA-DNA hybrid.
3. RNase H enzyme is used to remove poly (A) tails on mRNA.
Poly-A Polymerase:
The enzyme poIy-A polymerase plays a vital role in vitro gene manipulation techniques as it analyses the addition of AMP units to the 3′ end of RNA.
Essay # 12. Deoxyribonuclease I (Dnase I):
The enzyme DNase I is an endonuclease enzyme which digests either single or double-stranded DNA, producing a mixture of mononucleotides and oligonucleotides. DNase I hydrolyses each strand of double-stranded DNA independently and at random. Addition of Mg2+ to reaction mixture ensures random cleavage while addition of Mn2+ gives cleavage nearly at the same place on both strands. DNase enzyme is obtained mostly from bovine pancreas.
Uses of DNase I Enzyme:
DNase 1 enzyme is useful for a variety of applications including nick translation, DNA foot printing, bisulphite mediated mutagenesis and RNA purification.
Essay # 13. Terminal Deoxynucleotidyl Transferase Enzyme:
The enzyme deoxynucleotidyl transferase catalyses the repetitive addition of monodeoxynucleotide units from a deoxynucleoside triphosphate to the terminal 3′-hydroxyl group of a DNA molecule. This enzyme has a molecular weight of 32000 and consists of two subunits each with a molecular weight of 26500 and 8000. This enzyme is isolated from calf thymus.
Uses of Terminal Transferase Enzyme:
1. The enzyme terminal transferase is used to add homopolymer tails of DNA fragmeirts. Using a technique called homopolymer tailing, sticky ends can be built up on blunt-ended DNA molecules.
For examples, one preparation of DNA could be treated with the enzyme terminal transferase in the presence of dATP, resulting in the addition of a poly (dA) chain to each DNA strand. There is another preparation of DNA which provides 3 tails of poly (T) using same enzyme with TTP.
When both types of DNA preparations DNA fragments with poly A tails and DNA fragments with poly T tails, are mixed, there takes place base pairing between complementary sticky ends, which could then be ligated. One advantage of this method is that ligation does not take place between fragments from the same DNA preparation.
2. Terminal transferase enzyme is used for 3′-end labelling of DNA fragments
3. Terminal transferase enzyme is also used for the addition of single nucleotides to the 3- end of DNA for in vitro mutagenesis.
Purine and pyrimidine polymerization rates, by using terminal transferase enzyme, depend on the addition of Mg2+, Mn2+ or Co2+ in the reaction mixture.
Essay # 14. Reverse Transcriptase Enzyme:
The enzyme reverse transcriptase is isolated from avian myeloblastos virus (AMV). It is an RNA-dependent DNA polymerase. The enzyme requires DNA primer complementary to the RNA template, as well as presence of Mg2+ or Mn2+ for initiation of transcription. Reverse transcriptase mediates the conversion of genetic information present in single- stranded molecule of RNA into a double-stranded molecule of DNA.
Until recently, it was known that the genetic information’s of DNA pass to protein through mRNA. During 1960s, Temin and coworkers postulated that in certain cancer causing animal viruses which contain RNA as genetic material, transcription of cancerous genes (on RNA into DNA) takes places most probably by DNA polymerase directed by viral RNA.
Then DNA is used as template for synthesis of many copies of viral RNA in a cell. In 1970, S. Mizutani, H.M. Temin and D. Baltimore discovered that information can pass back from RNA to DNA.
They found that retroviruses (possessing RNA) contain RNA dependent DNA polymerase which IS also called reverse transcriptase. This process produces single- stranded DNA which in turn functions as template for complementary chain of DNA.
Reverse transcriptase enzyme has two subunits. The enzymatically active forms of the purified enzyme are α, β and αβ. The molecular weight of the α-subunit is 68000 and that of β-subunit is 92,000.
The mature α-β form is the most active form of AMV reverse transcriptase enzyme. It has several enzymatic roles such as RNA-directed DNA polymerase action, DNA dependent RNA polymerase activity and RNase-H activity.
The α- subunit of reverse transcriptase contains the polymerase activity. It also has the RNase- H activity during which degradation of RNA in DNA: RNA hybrids takes place. Such a sort of exonucleolytic activity of RNase-H enzyme can proceed either from the 5′- or 3′- terminus.
Uses of Reverse Transcriptase Enzyme:
1. The in vitro synthesis of cDNA from mRNA and other RNA molecule using reverse transcriptase enzyme has become a very important technique in the field of molecular biology.
2. DNA-dependent DNA polymerase activity of reverse transcriptase enzyme is responsible for second-strand formation in cDNA synthesis. Such a polymerising activity of reverse transcriptase is inhibited by the addition of actinomycin-D.
3. The reverse transcriptase enzyme mediates the conversion of genetic information present in single-stranded molecule of RNA into a double-stranded molecule of DNA.