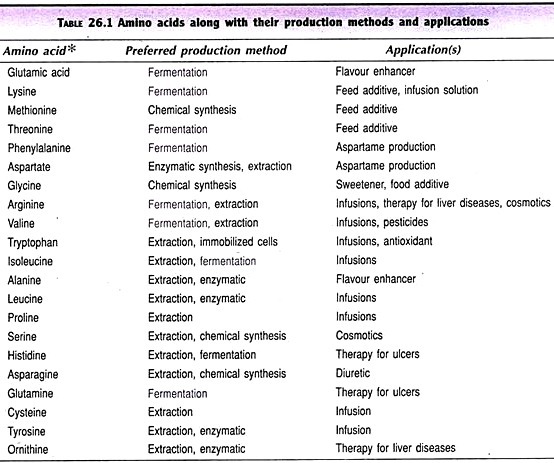
This article throws light upon the three principles of identification of DNA sequence. The three principles are: (1) Nucleic Acid Hybridization (2) DNA Probes and (3) DNA Chip-Microarray of Gene Probes.
Contents
Principle # 1. Nucleic Acid Hybridization:
Hybridization of nucleic acids (particularly DNA) is the basis for reliable DNA analysis. Hybridization is based on the principle that a single-stranded DNA molecule recognizes and specifically binds to a complementary DNA strand amid a mixture of other DNA strands. This is comparable to a specific key and lock relationship. The general procedure adopted for nucleic acid hybridization is as follows (Fig. 14.1).
The single-stranded target DNA is bound to a membrane support. Now the DNA probe (single- stranded and labeled with a detector substance) is added. Under appropriate conditions (temperature, ionic strength), the DNA probe pairs with the complementary target DNA.
The unbound DNA probe is removed. Sequence of nucleotides in the target DNA can be identified from the known sequence of DNA probe. There are two types of DNA hybridization-radioactive and non-radioactive respectively using DNA probes labeled with isotopes and non-isotopes as detectors.
Principle # 2. DNA Probes:
A DNA probe or a gene probe is a synthetic, single-stranded DNA molecule that can recognize and specifically bind to a target DNA (by complementary base pairing) in a mixture of biomolecules. DNA probes are either long (> 100 nucleotides) or short (< 50 nucleotides), and may bind to the total or a small portion of the target DNA. There is a wide variation in the size of DNA probes used (may range from 10 bases to 10,000 bases). The most important requirement is their specific and stable binding with target DNAs.
Methods Employed to Obtain DNA Probes:
A great majority of DNA probes are chemically synthesized in the laboratory. There are, however, many other ways of obtaining them-isolation of selected regions of genes, cloning of intact genes, producing from mRNAs.
Isolation of selected regions of genes:
The DNA from an organism (say a pathogen) can be cut by using restriction endonucleases. These DNA fragments are cloned in vectors and the DNA probes can be selected by screening.
Synthesis of DNA probes from mRNA:
The mRNA molecules specific to a particular DNA sequence (encoding a protein) are isolated. By using the enzyme reverse transcriptase, complementary DNA (cDNA) molecules are synthesized. This cDNA can be used as a probe to detect the target DNA.
Mechanism of Action of DNA Probes:
The basic principle of DNA probes is based on the denaturation and renaturation (hybridization) of DNA. When a double-stranded DNA molecule is subjected to physical (temperature > 95°C or pH < 10.5) or chemical (addition of urea or formaldehyde) changes, the hydrogen bonds break and the complementary strands get separated.
This process is called denaturation. Under suitable conditions (i.e., temperature, pH, salt concentration), the two separated single DNA strands can reassemble to form the original double-stranded DNA, and this phenomenon is referred to as renaturation or hybridization.
Radioactive Detection System:
The DNA probe is usually tagged with a radioactive isotope (commonly phosphorus-32). The target DNA is purified and denatured, and mixed with DNA probe. The isotope labeled DNA molecules specifically hybridizes with the target DNA (Fig. 14.1).
The non-hybridized probe DNA is washed away. The presence of radioactivity in the hybridized DNA can be detected by autoradiography. This reveals the presence of any bound (hybridized) probe molecules and thus the complementary DNA sequences in the target DNA.
Non-radioactive Detection System:
The disadvantage with the use of radioactive label is that the isotopes have short half-lives and involve risks in handling, besides requiring special laboratory equipment. So, non-radioactive detection systems (e.g., biotinylation) have also been developed. Biotin-labeled (biotinylated) nucleotides are incorporated into the DNA probe. The detection system is based on the enzymatic conversion of a chromogenic (colour producing) or chemiluminescent (light emitting) substrates.
The procedure commonly adopted for chemiluminescent detection of target DNA is depicted in Fig. 14.2. A biotin labeled DNA probe is hybridized to the target DNA. The egg white protein avidin or its bacterial analog streptavidin is added to bind to biotin. Now a biotin labeled enzyme, such as alkaline phosphatase is added which attaches to avidin or streptavidin. These proteins have four separate biotin-binding sites.
Thus, a single molecule (avidin or streptavidin) can bind to biotin-labeled DNA probe as well as biotin-labeled enzyme. On the addition of a chemiluminescent substrate, the enzyme alkaline phosphatase acts and converts it to a light emitting product which can be measured.
Advantages:
The biotin-labeled DNA is quite stable at room temperature for about one year. The detection devices using chemiluminescence are preferred, since they are as sensitive as radioisotope detection, and more sensitive than the use of chromogenic detection systems.
PCR in the use of DNA probes:
DNA probes can be successfully used for the identification of target DNAs from various samples — blood, urine, feces, tissues, throat washings without much purification. The detection of target sequence becomes quite difficult if the quantity of DNA is very low. In such a case, the polymerase chain reaction (PCR) is first employed to amplify the minute quantities of target DNA and identified by a DNA probe.
DNA Probes and Signal Amplification:
Signal amplification is an alternative to PCR for the identification of minute quantities of DNA by using DNA probes. In case of PCR, target DNA is amplified, while in signal amplification it is the target DNA bound to DNA probe that is amplified.
There are two general methods to achieve signal amplification.
1. Separate the DNA target—DNA probe complex from the rest of the DNA molecules, and then amplify it.
2. Amplify the DNA probe (bound to target DNA) by using a second probe. The RNA complementary to the DNA probe can serve as the second probe. The RNA-DNA-DNA complex can be separated and amplified. The enzyme O-beta replicase which catalyses RNA replication is commonly used.
Principle # 3. The DNA Chip-Microarray of Gene Probes:
The DNA chip or Gene-chip contains thousands of DNA probes (4000,000 or even more) arranged on a small glass slide of the size of a postage stamp. By this recent and advanced approach, thousands of target DNA molecules can be scanned simultaneously.
Technique for Use of DNA Chip:
The unknown DNA molecules are cut into fragments by restriction endonucleases. Fluorescent markers are attached to these DNA fragments. They are allowed to react to the probes of the DNA chip. Target DNA fragments with complementary sequences bind to DNA probes. The remaining DNA fragments are washed away. The target DNA pieces can be identified by their fluorescence emission by passing a laser beam. A computer is used to record the pattern of fluorescence emission and DNA identification.
The technique of employing DNA chips is very rapid, besides being sensitive and specific for the identification of several DNA fragments simultaneously. Scientists are trying to develop Gene-chips for the entire genome of an organism.
Applications of DNA Chip:
The presence mutations in a DNA sequence can be conveniently identified. In fact, Gene-chip probe array has been successfully used for the detection of mutations in the p53 and BRCA I genes. Both these genes are involved in cancer.