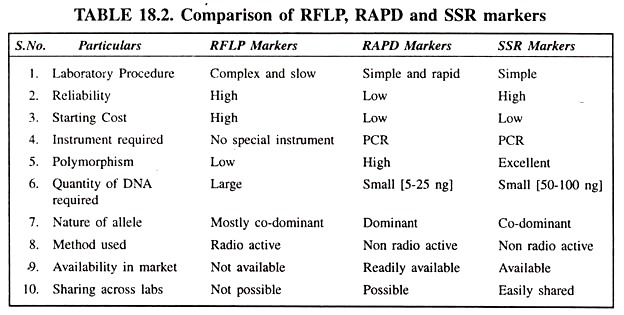
The following points highlight the top ten types of DNA markers. The types are: 1. Restriction Fragment Length Polymorphism 2. Amplified Fragment Length Polymorphism 3. Random Amplified Polymorphic 4. Cleaved Amplified Polymorphic Sequences 5. Simple Sequence Repeat (SSR) Length Polymorphism 6. Heteroduplex Analysis 7. Single Nucleotide Polymorphism 8. Single Nucleotide Polymorphism and Others.
Contents
Types of DNA Markers:
- Restriction Fragment Length Polymorphism (RFLP)
- Amplified Fragment Length Polymorphism (AFLP)
- Random Amplified Polymorphic DNA (RAPD)
- Cleaved Amplified Polymorphic Sequences (CAPS)
- Simple Sequence Repeat (SSR) Length Polymorphism
- Single Strand Conformational Polymorphism (SSCP)
- Heteroduplex Analysis (HA)
- Single Nucleotide Polymorphism (SNP)
- Expressed Sequence Tags (EST)
- Sequence Tagged Sites (STS)
DNA Marker: Type # 1.
Restriction Fragment Length Polymorphisms (RFLPs):
RFLPs refer to variations found within a species in the length of DNA fragments generated by specific endonuclease. RFLPs are first type of DNA markers developed to distinguish individuals at the DNA level. RFLP technique was developed before the discovery of Polymerase Chain Reaction (PCR).
The advantages, disadvantages and uses of this technique are presented below:
Advantages:
RFLP technique has several advantages. It is a cheaper and simple technique of DNA sequencing. It does not require special instrumentation. The majority of RFLP markers are co-dominant and highly locus specific. These are powerful tools for comparative and synteny mapping.
It is useful in developing other markers such as CAPS and INDEL. Several samples can be screened simultaneously by this technique using different probes. RFLP genotypes for single copy or low copy number genes can be easily scored and interpreted.
Disadvantages:
Developing sets of RFLP probes and markers is labour intensive. This technique requires large amount of high quality DNA. The multiplex ratio is low, typically one per gel. The genotyping throughput is low. It involves use of radioactive chemicals. RFLP finger prints for multi-gene families are often complex and difficult to score. RFLP probes cannot be shared between laboratories.
Uses:
They can be used in determining paternity cases. In criminal cases, they can be used in determining source of DNA sample. They can be used to determine the disease status of an individual. They are useful in gene mapping, germplasm characterization and marker assisted selection. They are useful in detection of pathogen in plants even if it is in latent stage.
DNA Marker: Type #
2.
Amplified Fragment Length Polymorphism (AFLP):
AFLPs are differences in restriction fragment lengths caused by SNPs or INDELs that create or abolish restriction endonuclease recognition sites. AFLP assays are performed by selectively amplifying a pool of restriction fragments using PCR. RFLP technique was originally known as selective restriction fragment amplification.
Advantages:
It provides very high multiplex ratio and genotyping throughput. These are highly reproducible across laboratories. No marker development work is needed; however, AFLP primer screening is often necessary to identify optimal primer specificities and combinations.
No special instrumentation is needed for performing AFLP assays; however, special instrumentation is needed for co-dominant scoring.
Start-up costs are moderately low. AFLP assays can be performed using very small DNA samples (typically 0.2 to 2.5 pg per individual). The technology can be applied to virtually any organism with minimal initial development.
Disadvantages:
The maximum polymorphic information content for any bi-allelic marker is 0.5. High quality DNA is needed to ensure complete restriction enzyme digestion. DNA quality may or may not be a weakness depending on the species. Rapid methods for isolating DNA may not produce sufficiently clean template DNA for AFLP analysis.
Proprietary technology is needed to score heterozygotes and ++ homozygotes. Otherwise, AFLPs must be dominantly scored. Dominance may or may not be a weakness depending on the application.
The homology of a restriction fragment cannot be unequivocally ascertained across genotypes or mapping populations. Developing locus specific markers from individual fragments can be difficult and does not seem to be widely done.
The switch to non-radioactive assays has not been rapid. Chemiluminescent AFLP fingerprinting methods have been developed and seem to work well.
The fingerprints produced by fluorescent AFLP assay methods are often difficult to interpret and score and thus do not seem to be widely used. AFLP markers often densely cluster in centromeric regions in species with large genomes, e.g., barley (Hordeum vulgare L.) and sunflower (Helianthus annuus L.).
Uses:
This technique has been widely used in the construction of genetic maps containing high densities of DNA marker. In plant breeding and genetics, AFLP markers are used in varietal identification, germplasm characterization, gene tagging and marker assisted selection.
DNA Marker: Type #
3.
Random Amplified Polymorphic DNA (RAPDs):
RAPD refers to polymorphism found within a species in the randomly amplified DNA generated by restriction endonuclease enzyme. RAPDs are PCR based DNA markers. RAPD marker assays are performed using single DNA primer of arbitrary sequence.
Advantages:
RAPD primers are readily available being universal. They provide moderately high genotyping throughput. This technique is simple PCR assay (no blotting and no radioactivity). It does not require special equipment. Only PCR is needed. The start-up cost is low.
RAPD marker assays can be performed using very small DNA samples (5 to 25 ng per sample). RAPD primers are universal and can be commercially purchased. RAPD markers can be easily shared between laboratories. Locus-specific, co-dominant PCR-based markers can be developed from RAPD markers. It provides more polymorphism than RFLPs.
Disadvantages:
The detection of polymorphism is limited. The maximum polymorphic information content for any bi-allelic marker is 0.5. This technique only detects dominant markers. The reproducibility of RAPD assays across laboratories is often low. The homology of fragments across genotypes cannot be ascertained without mapping. It is not applicable in marker assisted breeding programme.
Uses:
This technique can be used in various ways such as for varietal identification, DNA fingerprinting, gene tagging and construction of linkage maps. It can also be used to study phylogenetic relationship among species and sub-species and assessment of variability in breeding populations.
DNA Marker: Type #
4.
Cleaved Amplified Polymorphic Sequences (CAPS):
CAPS polymorphisms are differences in restriction fragment lengths caused by SNPs or INDELs that create or abolish restriction endonuclease recognition sites in PCR amplicons produced by locus-specific oligonucleotide primers.
CAPS assays are performed by digesting locus-specific PCR amplicons with one or more restriction enzymes and separating the digested DNA on agarose or polyacrylamide gels.
CAPS analysis is versatile and can be combined with single strand conformational polymorphim (SSCP), sequence-characterized amplified region (SCAR), or random amplified polymorphic DNA (RAPD) analysis to increase the chance of finding a DNA polymorphism.
Michaels and Amasino (1998) proposed a variant of the CAPS method called dCAPS based on SNPs.
Advantages:
The genotyping throughput is moderately high. It is a simple PCR assay. Markers are developed from the DNA sequences of previously mapped RFLP markers. Most CAPS markers are co- dominant and locus specific. No special equipment is needed to perform manual CAPS marker assays.
CAPS marker assays can be performed using semi-automated methods, e.g., fluorescent assays on a DNA sequencer (e.g., ABI377). Start-up costs are low for manual assay methods. CAPS assays can be performed using very small DNA samples (typically 50 to 100 ng per individual). Most CAPS genotypes are easily scored and interpreted. CAPS markers are easily shared between laboratories.
Disadvantages:
Typically, a battery of restriction enzymes must be tested to find polymorphisms. Although CAPS markers still nave great utility and should not be over looked, other methods have emerged as tools for screening locus-specific DNA fragments for polymorphisms, e.g., SNP assays. The development of easily scored and interpreted assays may be difficult for some genes, especially those belonging to multi-gene families.
Uses:
This is straightforward way to develop PCR-based markers from the DNA sequences of previously mapped RFLP markers. It is a simple method that builds on the investment of an RFLP map and eliminates the need for DNA blotting.
DNA Marker: Type #
5.
Simple Sequence Repeats (SSRs):
Simple sequence repeats (SSRs) or microsatellites are tandemly repeated mono-, di-, tri-, tetra-, penta-, and hexanucleotide motifs. SSR length polymorphisms are caused by differences in the number of repeats. SSR loci are individually amplified by PCR using pairs of oligonucleotide primers specific to unique DNA sequences flanking the SSR sequence.
Jeffreys (1985) showed that some restriction fragment length polymorphisms are caused by VNTRs. The name “mini satellite” was coined because of the similarity of VNTRs to larger satellite DNA repeats.
Advantages:
SSR markers tend to be highly polymorphic. The genotyping throughput is high. This is a simple PCR assay. Many SSR markers are multi-allelic and highly polymorphic. SSR markers can be multiplexed, either functionally by pooling independent PCR products or by true multiplex- PCR. Semi-automated SSR genotyping methods have been developed. Most SSRs are co-dominant and locus specific.
No special equipment is needed for performing SSRs assays; however, special equipment is needed for some assay methods, e.g., semi-automated fluorescent assays performed on a DNA sequences. Start-up costs are low for manual assay methods (once the markers are developed). SSR assays can be performed using very small DNA samples (~100 ng per individual). SSR markers are easily shared between laboratories.
Disadvantages:
The development of SSRs is labor intensive. SSR marker development costs are very high. SSR markers are taxa specific. Start-up costs are high for automated SSR assay methods. Developing PCR multiplexes is difficult and expensive. Some markers may not multiplex.
Uses:
SSR markers are used for mapping of genes in eukaryotes.
DNA Marker: Type #
6.
Single Strand Conformational Polymorphisms (SSCPs):
SSCPs refer to DNA polymorphisms produced by differential folding of single-stranded DNA harboring mutations. The conformation of the folded DNA molecule is produced by intra-molecular interactions and is thus a function of the DNA sequence.
SSCP marker assays are performed using heat-denatured DNA on non-denaturing DNA sequencing gels. Special gels (e.g., mutation detection enhancement gels) have been developed to enhance the discovery of single-strand conformational polymorphisms caused by INDELs, SNPs, or SSRs.
Advantages:
It is a simple PCR assay. Many SSCP markers are multi-allelic and highly polymorphic. Most SSCPs are co-dominant and locus specific. No special equipment is needed. Start-up costs are low. SSCP marker assays can be performed using very small DNA samples (typically 10 to 50 ng per individual).
SSCP markers are easily shared between laboratories. SSCP gels can be silver stained (no radioactivity). The complexity of PCR products can be assessed and individual fragments can be isolated and sequenced.
Disadvantages:
The development of SSCP markers is labor intensive. SSCP marker development costs can be high. SSCP marker analysis cannot be automated.
Uses:
SSCPs have been widely used in human genetics to screen disease genes for DNA polymorphisms. Although SSCP analysis does not uncover every DNA sequence polymorphism, the methodology is straight forward and a significant number of polymorphisms can be discovered. SSCP analysis can be a powerful tool for assessing the complexity of PCR products.
DNA Marker: Type #
7.
Heteroduplex Analysis (HA):
It refers to DNA polymorphisms produced by separating homo-duplex from heteroduplex DNA using non-denaturing gel electrophoresis or partially denaturing high performance liquid chromatography.
Single-base mismatches between genotypes produce hetero-duplexes; thus, the presence of hetero-duplexes signals the presence of DNA polymorphisms. Heteroduplex analyses can be rapidly and efficiently performed on numerous genotypes before specific alleles are sequenced, thereby greatly reducing sequencing costs in SNP discovery and SNP marker development.
Advantages:
It is a powerful method for SNP discovery. Automated HA can be performed using HPLC. Most heteroduplex markers are co-dominant and locus specific. HA can be performed using very small DNA samples (typically 10 to 50 ng per individual). HA markers are easily shared between laboratories.
Disadvantages:
Requires special equipment. One protocol may not be sufficient for heteroduplex analyses of different targets via HPLC.
Uses:
Heteroduplex analysis has been mostly used in human genetics to screen disease genes for DNA polymorphism. In plant breeding, it is used for detection of pathogens which are in latent stage and thus useful in selection of disease free plants. It is also useful in the discovery of single nucleotide polymorphism.
DNA Marker: Type #
8.
Single Nucleotide Polymorphism (SNP):
The variations which are found at a single nucleotide position are known as single nucleotide polymorphisms or SNP. Such variation results due to substitution, deletion or insertion. This type of polymorphisms has two alleles and also called bialleleic loci. This is the most common class of DNA polymorphism. It is found both in natural lines and after induced mutagenesis. Main features of SNP markers are given below.
1. SNP markers are highly polymorphic and mostly bialleleic.
2. The genotyping throughput is very high.
3. SNP markers are locus specific.
4. Such variation results due to substitution, deletion or insertion.
5. SNP markers are excellent long term investment.
6. SNP markers can be used to pinpoint functional polymorphism.
7. This technique requires small amount of DNA.
Advantages:
SNP markers are useful in gene mapping. SNPs help in detection of mutations at molecular level. SNP markers are useful in positional cloning of a mutant locus. SNP markers are useful in detection of disease causing genes.
Disadvantages:
Most of the SNPs are bialleleic and less informative than SSRs. Multiplexing is not possible for all loci. Some SNP assay techniques are costly. Development of SNP markers is labour oriented. More (three times) SNPs are required in preparing genetic maps than SSR markers.
Uses:
SNPs are useful in preparing genetic maps. They have been used in preparing human genetic maps. In plant breeding, SNPs have been used to lesser extent.
DNA Marker: Type #
9.
Expressed Sequence Tags (EST):
Expressed Sequence Tags (ESTs) are small pieces of DNA and their location and sequence on the chromosome are known. The variations which are found at a single nucleotide position are known. The term Expressed Sequence Tags (ESTs) was first used by Venter and his colleagues in 1991. Main features of EST markers are given below.
1. ESTs are short DNA sequences (200-500 nucleotide long).
2. They are a type of sequence tagged sites (STS).
3. ESTs consist of exons only.
Advantages:
It is a rapid and inexpensive technique of locating a gene. ESTs are useful in discovering new genes related to genetic diseases. They can be used for tissue specific gene expression.
Disadvantages:
ESTs have lack of prime specificity. It is a time consuming and labour oriented technique. The precision is lesser than other techniques. It is difficult to obtain large (> 6kb) transcripts. Multiplexing is not possible for all loci.
Uses:
ESTs are commonly used to map genes of known function. They are also used for phylogenetic studies and generating DNA arrays.
DNA Marker: Type #
10.
Sequence Tagged Sites (STS):
In genomics, a sequence tagged site (STS) is a short DNA sequence that has a single copy in a genome and whose location and base sequence are known. Main features of STS markers are given below.
1. STSs are short DNA sequences (200-500 nucleotide long).
2. STSs occur only once in the genome.
3. STS are detected by PCR in the presence of all other genomic sequences.
4. STSs are derived from cDNAs.
Advantages:
STSs are useful in physical mapping of genes. This technique permits sharing of data across the laboratories. It is a rapid and most specific technique than DNA hybridization techniques. It has high degree of accuracy. It can be automated.
Disadvantages:
Development of STS is a difficult task. It is time consuming and labour oriented technique. It require high technical skill.
Uses:
STS is the most powerful physical mapping technique. It can be used to identify any locus on the chromosome. STSs are used as standard markers to find out gene in any region of the genome. It is used for constructing detailed maps of large genomes.