Some of the major cell organs involved in ultra-structure of prokaryotic cell are as follows:
1. Cell envelope 2. Cytoplasm 3. Nucleoid 4. Appendages.
Prokaryotic cells are the simplest of most primitive cells. The records of microfossils suggest that they have evolved 2.5 billion years ago and existed as the only organisms on earth for the next one billion years until eukaryotes evolved about 1.5 billion years ago. Earlier claims that oldest prokaryotic microfossils, the stromatolites (i.e., giant colonies of ancient cyanobacteria or BGA) of 3.5 billion years ago are actually lifeless mineral artifacts.
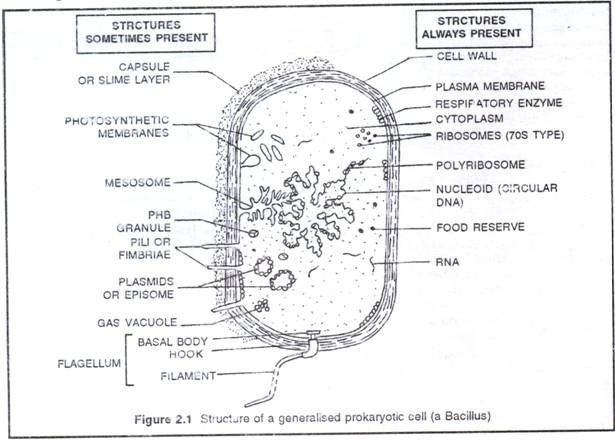
The Prokaryotic ceil is the structural unit of two microbial groups: the archaebacteria and the eubacteria. Despite variations in shape and size, the fundamental structures of prokaryotic cells are same. Each prokaryotic cell is essentially a one envelop system that consists of protoplasm encased within cell envelope. The ultrastructure of a prokaryotic cell, particularly a typical bacterial cell consists of cell envelope, cytoplasm, nucleoid, plasmids and surface appendage.
1. Cell envelope:
It is the protective covering of bacterial cell that has three basic layers: the outermost glvcocalyx, middle cell wall and innermost cell membrane (plasma membrane),
(i) Clycocalyx:
It is the outermost layer of cell envelope which chemically composed of polysaccharides with or without proteins. When glycocalyxis thick and tough, it is called capsule, and when it forms a loose sheath it is called slime layer.
Though not essential for bacterial survival, glycocalyx has many functions:
(a) Protects cell from desiccation, toxins and phagocytes,
(b) Helps in adhesion, immunogenicity and virulence.
(ii) Cell wall:
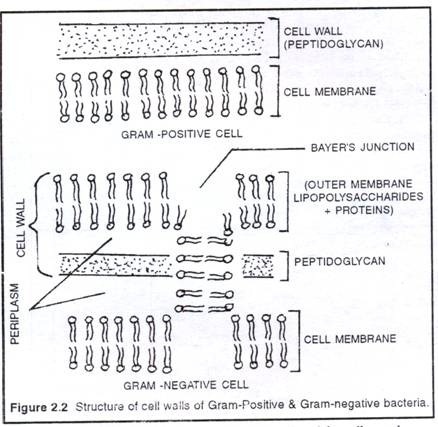
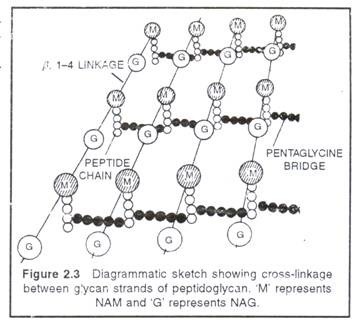
It is the rigid middle layer of cell envelope that provides shape and prevents a bacterium from osmotic bursting in a hypotonic solution. In Gram-positive bacteria, cell wall is single layered and almost uniform in thickness (10 to 80nm).It is composed of peptidoglycan (murein or mucopeptide), which consists of a three dimensional network of glyean strands cross linked by peptide chains. Each glycan strand is 20-130 units long consists of two alternating amino sugars, N-acetylglucosamine (NAG) and N-acetylmuramicacid (NAM).
Certain antibiotics (penicillin) lysozyme prevents cross linking and kills bacteria. The walls of Gram- positive bacteria also contain teichoic acids (Polyphosphate Polymery but proteins are almost absent. Teichoic acids act as surface antigen. In Gram-negative bacteria cell wall is two- layered and only 7.5-12 nm thick. The inner layer in a thin peptidoglycan and the outer layer is a unit membrane called outer membrane. The outer membrane is a lipid bilayer consists of phospholipids, proteins and a unique lipid, lipopolysaccharide (LPS).
The LPS molecules present only in the outer face of the outer membrane. The outer membrane also contains channel like proteins called porins, through which low molecular weight substances enter or exit. In some Gram-negative bacteria (.Mycobacterium, Noccardia) the wall contains long chain fatty acids called mycolic acids. In the walls of Gram- negative bacteria, a fluid- filled space present on either side of the peptidoglycan layer i.e. between the outer membrane of cell wall and the cell membrane. This space is called periplasm or periplasmic gel. It acts like bacterial lysosome.
Many species of bacteria are wall-less, either develop spontaneously or induced by cell wall degrading agents. These bacteria are called L-forms, after the Lister Institute, London. The L- forms of bacteria may develop cell wall but mollicutes will never develop cell wall.
(iii) Cell membrane:
It is the innermost layer of cell envelope. It is a semi-permeable, quasi-fluid, dynamic membrane similar to that of eukaryotic membrane. But the only difference is that, in bacteria they lack sterols instead hopanoids present. The hopanoids are pentacyclic sterol-like molecules that stabilize the bacterial cell membrane. In Gram-negative bacteria at certain places, the outer face of plasma membrane is continuous with the inner face of the outer membrane to form Bayer’s junctions. There are about 200-400 Layer’s junctions present in a Gram- negative cell.
In a bacterial cell, plasma membrane performs many functions:
(a) It retains the cytoplasm
(b) Prevent loss of essential components through leakage
(c) Aids in the movement of nutrients, wastes and secretions across the membrane
(d) Holds receptor molecules that detect and respond chemicals in their surroundings. Such as respiration, photosynthesis, synthesis of lipids and cell wall constituents
(f) It invaginates to form mesosome and thylakoids of cyanobacteria.
2. Cytoplasm:
It is granular, crystallo-colloidal complex that fills the whole prokaryotic cell excluding nucleoid. The cytoplasm contains mesosome, chromatophores, ribosomes, inclusion bodies and plasmids.
(i) Mesosome (or chondrioids):
It is a convoluted membranous infolding of the plasma membrane. Mesosome when connected with nucleoid is called septal mesosome and when not connected called lateral mesosome.
Mesosome helps in:
(a) Cell wall formation
(b) Chromosome replication and distribution to daughter cells
(c) Secretory activity
(d) Increase the surface area of plasma membrane
(e) Mesosome can be considered analogous to mitochondria of eukaryotic cells as these are the sites of respiratory enzymes. Hence, mososomes are also called as “mitochondria of prokaryotic cells” or “bacterial mitochondria”.
(ii) Chromatophores:
These are internal membrane systems of prokaryotic cells. These are very extensive and complex in photosynthetic forms like cyanobacteria and purple bacteria where they are called as thylakoids. In nitrifying bacteria the chromatophores increase metabolic area.
(iii) Ribosomes:
Prokaryotic ribosomes are 70S in nature that consist of larger 5OS and smaller 30S subunits. During protein synthesis, about 4-8 ribosomes attach to a single mRNA to form polyribosomes or polysomes. Non functional ribosomes present in separated subunits.
(iv) Inclusion bodies:
These are non-living structures present freely in the cytoplasm. Inclusion bodies may be organic or inorganic. They include mainly food reserve and special prokaryotic organelles like gas vacuoles, chromosomes, carboxysomes, and magnetosomes. Except food reserve other inclusion bodies are surrounded by a single layer non-unit membrane which is 2-5 nm thick.
Food reserve:
These are reserve materials or storage granules which are not bounded by any membrane system. Generally, a given bacterial species store only one kind of reserve material. Further, the cellular content of reserve material is low in actively growing cells but them increase when cells are short of nitrogen.
The volutin granules (= polyphosphate granules) and sulphur granules are inorganic inclusion bodies which store phosphate and sulphur respectively. These granules are also called meta-chromatic granules because of their ability to take different colours to basic dyes. The organic- food reserves present in some bacteria are glycogen granules, protein granules (=cyanophycin), cyanophycean granules (= starch granules) and PHB granules. The PHB (Poly β-hydroxybutyrate) granules are storage reservoir of fatty acids in case of Pseudomonas, Bacillus, Azotobacter species. The PHB is commercially used to prepare biodegradable plastics.
Gas vacuoles:
These are the organic inclusions of most aquatic, free floating forms. Each gas vacuole is an aggregate of variable number of hollow, cylindrical gas vesicles. Gas vacuoles help in floating, for proper positioning in water to trap sunlight for photosynthesis and protect against harmful radiations.
Chlorosomes:
These are cigar-shaped vesicles that enclose photosynthetic pigments like bacteriochlorophyll c, d, or e. Chlorosomes are distinct structures found just below the plasma membrane but tightly joint to it by a basal plate. These are found in the green bacteria.
Carboxysomes:
These are principal sites of CO2 fixation in case of autotrophic prokaryotes like cyanobacteria, purple bacteria, nitrifying bacteria etc.
Magnetosomes:
These are the vesicles filled with crystals of magnetite (Fe3O4). Magneto-somes help the bacteria to orient themselves in a magnetic field and determine the direction of swimming.
(v) Plasmids:
Laderberg and Hays (1952) introduced the term ‘plasmid’ to those ring-like self replicating extra chromosomal double stranded DNA that are found in the cytoplasm of prokaryotes. They are also found in eukaryotes (yeast) and their organelles. Plasmids are used as in ideal vector for indirect gene transfer in recombinant DNA technology (Genetic Engineering).
Plasmids are generally double stranded closed circles of D.N A with sizes vary from 1-3 00 kilobase pairs (1 Kbp = 1000bp) and carries 5-100 genes. Hence, plasmids are often called as minichromosomes. They are also known as dispensable autonomous elements because the genes they carry have no role in viability and bacterial growth.
The average number of plasmids per bacterial cell is called copy number. Plasmids with low copy number (1-2) are called single copy plasmids, while those with high copy number (10-30) are called multi-copy plasmids. Some plasmids can temporarily integrate or can detach from main chromosome and are called as episomes, e.g., F-plasmid. It is to be noted that all episomes are plasmids but all plasmids are not episomes. The term episome was coined by Jacob and Wollman 1958).
Plasmid Types:
On the basis of function, plasmids are of following types –
(a) Sex plasmid:
It carries fertility factor (F-factor) responsible for the formation of sex-pili and conjugation. Hence, often called F-plasmid.
(b) R-Plasmid:
It carries resistance (R) factor which provide resistance again si antibiotics, heavy metals, UV-radiation etc., e.g. Rl.R4G etc.
(c) Col-plasmid:
It carries colicinogenic factor that, produce colicins (bacteriocins) to kill other bacterial.
(d) Degradative plasmid:
It decompose hydracarbon in petroleum, e g. present in Pseudomonas putida [A genetically engineered bacterium which would degrade all the four types of substrates i.e. OCT (Octane, Hexane, Dcene), XYL (Xylene, Toluene), CAM (camphor) and NAH (naphthalene)}
(e) Ti-plasmids:
These are tumour including plasmids carried by the Agrabacterium tumefaciens. Ti-plasmid carries
T-DNA (transforming DNA) which is 200 Kbp long and causes crown gall disease in plants, T- DNA is an ideal vector for gene transfer in plants.
(f) Ri-Plasmids:
A subground of Ti-plasmids inducing the hairy root tumours e.g.,-A-rhizogene.
Plasmid can be categorised on the basis of number of copies per cell
(a) Released Plasmid: It normally maintain multiple copies per cell
(b) Stringent Plasmid: It has limited number of copies per cell
3. Nucleoid
Nucleoid is the genetic material of a prokaryotic cell that occupies up to 1/5 of the interior of the bacterial cell. It is represented by a single circular naked ds DNA which is highly looped and super coiled with the help of polyatnines (nucleoid proteins) and RNA. Nucleoid is a compact structure hat nuclear envelope and nucleolus, and therefore it is not an organized nucleus rather an incipent nucleus. In Escherichia coli the compact DNA is 1.2 mm in length which is about 250-700 times the length of the cell. The nucleoid is attached to the plasma membrane directly or by mesosomes.
4. Appendages:
The surface appendages present on bacterial cell may be motile flagellum or non-motile pili and fimbriae.
(a) Flagella (sing. Flagellum):
These are long (1-71m) fine hairy locomotary appendages present on bacterial surface for swimming. Their number and arrangement is called flagellation which is characteristic features of different genera of bacteria
Some bacteria bear sheath flagella surrounded by extension of cell membrane. In Vibrio, flagellation is mixed type where polar sheathed flagellum present along with many peritrichously arranged unsheathed flagella.
The ultrastructure of each flagellum shows 3 parts – basal body, hook and filament. The Basal body consists of a central root with one two pairs of rings. In Gram positive bacteria the basal body possesses two pairs of rings; the outer pair (L and P rings) remains attached to the outer membrane, whereas the inner pair (S and M rings) remains connected to the cell membrane. In Gram- positive bacteria, on the other hand, only the inner pair (Sand M) is present and the remains attached to the cell membrane (fig. 2.4).
The hook is slightly wider and curved structure about45 nm long that connects, basal body with the filament. The filament is hollow cylindrical structure about 1-70 nm long and 20 nm in diameter. Filament composed of 3-8 spiral of flagellian proteins. But hook is made up of a different kind of proteins.
The bacterial flageilum rotate by 360″ rather than a whip like back and forth movement. As a result the bacterial cell spins in the opposite direction and pushes the bacterium in forward direction.
(b) Pili (sing. Pilus) and fimbriae:
These two terms have often been interchangeably used. The pili are tubular outgrowths of about 18-20 nm made up of pilin protein. They are reported only in donor cell Gram negative bacteria where they help in conjugation. Hence, pili are also called sex-pili or F-pili. Their number is 1 -4 per cell. Fimbriae are bristle like surface appendages help in adhesion and mutual clinging length varies from 6.1 – 7.5 nm and diameter 3-10 nm.