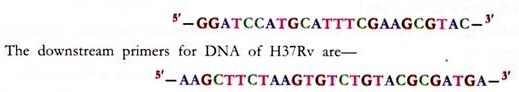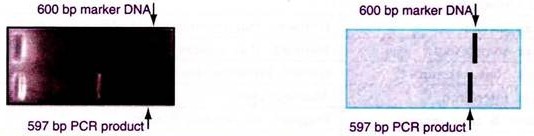The following points highlight the top seven applications of biotechnology. The applications are: 1. Nucleic Acid Probes 2.Gene Sequencing 3. Artificial Synthesis of DNA 4. Polymerase Chain Reaction 5. Subunit Vaccines 6. Organ, Tissue, Cell Culture and 7. Hybridoma Technique.
Contents
- Application of Biotechnology # 1. Nucleic Acid Probes:
- Application of Biotechnology # 2.Gene Sequencing:
- Application of Biotechnology # 3. Artificial Synthesis of DNA:
- Application of Biotechnology # 4. Polymerase Chain Reaction:
- Application of Biotechnology # 5. Subunit Vaccines:
- Application of Biotechnology # 6. Organ, Tissue, Cell Culture:
- Application of Biotechnology # 7. Hybridoma Technique:
Application of Biotechnology # 1. Nucleic Acid Probes:
Nucleic acid (NA) probes are pieces of nucleic acid (DNA or RNA) that can be used to identify the presence of a gene of interest. This probe is linked either to a radioactive substance, fluorescent compound, or an enzyme that gives a coloured product in order to be detected. The probe is bound if a sample of nucleic acid contains a base sequence complementary to that of the probe.
Uses of DNA Probes:
DNA probes are being used:
(a) To identify genes of interest to differentiate normal and mutated genes.
(b) To identify oncogenes in biopsy sample.
(c) To detect DNA polymorphism due to variation in a single locus caused by point mutation.
(d) Detection of gene causing genetic diseases and screen foetus for genetic disorders.
(e) To detect presence of pathogenic organisms in blood or tissue samples.
(f) As primers for PCR.
(g) Pathogens can be identified by detecting specific RNA base sequences with probes.
Methods for Preparation of DNA Probes:
Probes are synthesized by three methods viz.:
(1) Synthetic oligonucleotide probe
(2) PCR generated probes and
(3) In vivo cloning.
These methods are based upon the knowledge of the nucleotide sequence of many genes.
Application of Biotechnology # 2.Gene Sequencing:
The method for deducting the sequence of nucleotide bases in a gene or DNA is known as gene sequencing.
There are three methods for gene sequencing:
(a) Chemical cleavage method
(b) Enzymatic method and
(c) Oligonucleotide hybridization.
1. Chemical Cleavage Method or Maxam-Gilbert Method:
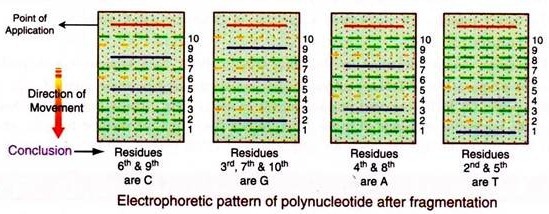
A DNA molecule can be cleaved chemically at a particular nucleotide base into small fragments. DNA can be cleaved specifically at G, A and G, C and T or only C, by adopting different chemical procedures. If DNA is heated at neutral pH, then the glycosidic bond (the bond between the sugar and base) of a methylated purine is broken off, when the heating is continued after adding alkali, the backbone of the fragment at G is broken. If this heating is done in dilute acid, the backbone at both A & G is broken.
Treatment of DNA with hydrazine in 2M NaCl and then with piperidine cleaves at C only. Treatment with only piperidine cleaves both at C & T. Using any one of these treatments, DNA is cleaved at a specific point. Supposing the following is the sequence of DNA whose sequencing is to be done
This labeled molecule is taken into four different tubes. Each tube is treated with different chemical procedures, so as to cleave either at A, T, G, or C, as described above. This results in the DNA molecule breaking into the following smaller fragments. (Fragments having the label are only considered)
Each of these four mixtures is separately subjected to electrophoresis on polyacrylamide gel (PAGE). In this electrophoresis the polynucleotide moves according to the number of nucleotide residues they contain, wherein the smallest fragment will move faster.
Difference in a single nucleotide from one fragment to the other results in a great difference in their migration fronts. Thus these fragments can be separated depending upon their number of nucleotides. The exact position of only the labelled (32P) fragments in the gel can be determined by autoradiography on a photographic film.
The results of these procedures are compared with the calibrated electrophoretic pattern corresponding to nucleotides having from 1 to 10 bases.
Thus the electrophoretic pattern for the above polynucleotide after fragmentation with different chemical methods will be as follows:
It is seen that the labelled fragments obtained by deletion of C residues moved at rates indicate that they had 5 and 8 nucleotide units. Thus residues 6th and 9th of the original polynucleotide must have been C.
Likewise tallying the number of nucleotides in each set of the chemically reacted fragments derive that the sequence of the polynucleotide is:
To depict the sequence of the complete DNA molecule, it is fragmented by different restriction endonucleases and each of the resulting fragment is sequenced as above and the overlaps in the fragments helps in arranging the sequence of the complete DNA.
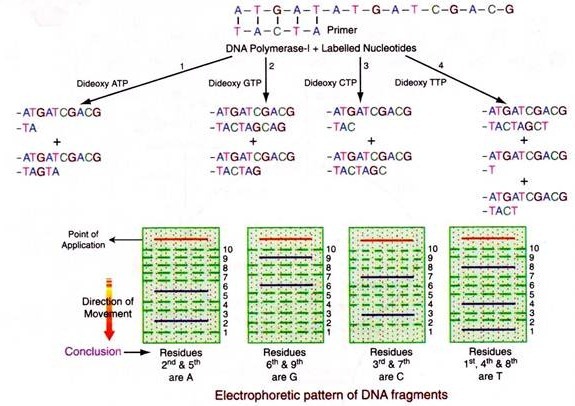
2. Enzymatic Method or Frederick Sanger Method:
Biologically DNA is duplicated by enzymes using each of the strands as a template i.e. a complementary strand is synthesized to each of the strands. The enzyme for replication is known as DNA polymerase (I, III). The single stranded DNA fragment, whose sequence is to be determined is taken in a tube and to it is added a short length of complimentary primer and the enzyme DNA polymerase-I (Klenow fragment), radioactively labelled four deoxyribonucleotides and any one of the 2′,3′-dideoxy nucleotide analog is taken.
The enzyme adds on nucleotide by nucleotide and whenever the analog is added, growth of the new chain is blocked because it lacks the 3′-hydroxyl end needed to form the next phosphodiester bond. Hence, fragments of various lengths are produced in which the dideoxy analog is at the 3′ end. Four such sets of chain terminated fragments (one of each dideoxy analog – A,T,G,C) are then electrophorised and the base sequence of the new DNA is read from the autoradiogram of the four lines.
The sequence is TACTAGCTG, which is the complementary strand.
Hence the original strand will contain the sequence:
Application of Biotechnology # 3. Artificial Synthesis of DNA:
DNA can be synthesized with a desired sequence of nucleotides, by the use of chemical reactions in the laboratory, which are fast, easy and inexpensive.
Procedure:
A chromatographic column is filled with controlled pore glass (CPG) beads, which acts as an inert solid support. The first nucleoside of the 3′-end is attached to a long chain alkyl amine-succinate which is the spacer linked to the glass bead. The 5′-OH group is protected with dimethoxytrityl (DMT) group. Further every nucleotide being added has a 5′-DMT group and a di-isopropyl amine group at 3′-phosphite group with a methyl group. This structure is called phosphoramidite.
Further the amino groups of all the bases are protected from cross reaction with benzoyl and isobutyl groups.
The nucleotide chain is elongated on the first nucleoside attached to glass beads through the spacer, in a cyclic manner as follows:
(a) The column is washed with acetonitrile to remove water and any nucleophiles present.
(b) Acetonitrile is then removed from the column by flushing it with argon.
(c) TCA is added to remove the 5′-DMT (detrityladon step) and thus produce a reactive 5′-OH group.
(d) To remove TCA, the column is washed with acetonitrile which is in turn flushed out with argon.
(e) The next base (phosphoramidite) and tetrazole are introduced into the column.
(f) Tetrazole activates the phosphoramidite to form a covalent bond between its 3′-phosphite and 5′-OH group of the first nucleoside (coupling step).
(g) Argon is flushed to remove unused phosphoramidite and tetrazole.
(h) The inter-nucleotide linkage in the form of phosphite triester bond is unstable and hence it is oxidized with iodine mixture to form phosphate triester bond (oxidation).
(i) Those support-bound nucleosides which are not coupled to the second nucleotide added are inactivated for further reaction by acetic anhydride and dimethyl-amino pyridine (capping step).
(j) The column is washed again to remove all the unwanted material.
(k) Then the cycles of detritylation, phosphoramidite activation, coupling, capping and oxidation are repeated till all the nucleotides are added. (1) When all the cycles are completed, the methyl groups of phosphate triester are removed with triethyl ammonium thiophenate in dioxane.
(m) The oligonucleotide is cleaved from the support by concentrated ammonia and eluted out of the column.
(n) The bases are de-protected (benzyl and isobutyl groups removed) by heating to 50°C in concentrated ammonia solution for 6 hours.
(o) The 5′-DMT is removed with 80% acetic acid.
(p) The 5′-terminal end of the artificially synthesized DNA strand is phosphorylated by an enzyme T4 polynucleotide kinase using ATP.
(q) The synthesized DNA is finally purified by HPLC.
Uses of Artificially Synthesized DNA by Chemical Method:
(1) Chemically synthesised DNA, with a specific sequence are used as primers for polymerase chain reaction.
(2) They are also used in DNA sequencing-as a short length of primer, complementary to the sequencing DNA fragment, in the enzymatic method of DNA sequencing.
(3) Oligonucleotides are used for creating mutations i.e. an oligonucleotide is synthesized with an altered base and is introduced in place of the original gene. This improves the performance of many bacteria or various human/animal/plant genetic diseases can be treated.
(4) If the amino acid sequence is known, then a DNA strand is synthesized using the codons for these amino acids. The gene synthesized for this protein is used for gene cloning.
(5) Double stranded DNA strands, called linkers can be synthesized containing restriction endonuclease recognition site, which helps in the cloning of DNA fragments (after breaking and joining).
(6) DNA sequences called adaptor sequences can be synthesized, which contains sites for two different restriction endonucleases-one used for insertion of the DNA and the other for excision at a later time.
(7) DNA fragments synthesized are used as DNA probes for diagnosis of various molecular diseases.
(8) Chemically synthesized DNA fragments are also used in the treatment of genetic disorders.
Application of Biotechnology # 4. Polymerase Chain Reaction:
Polymerase chain reaction is abbreviated as P.C.R. technology and termed as in vitro enzymatic gene amplification or in vitro gene cloning without its expression. It resembles the electronic Xeroxing of pages, hence this process can also be termed as ‘DNA Xeroxing’. P.C.R. technology is the amplification/cloning of DNA (or a gene) in a test-tube, using a mixture of template + primers + enzyme + bases (dNTPs) + buffer, by cycling the temperature within the reaction tube.
Procedure:
A micro test tube is taken with the DNA template (the sample), that is to be amplified, along with tris buffer (50 mM KCl + 10 mM tris HCl) to get a pH of 8.4. MgCl2 (1.5 mM) and 100 ng of gelatin are additional components required to stabilize the reaction mixture.
Sufficient concentration (200 µM each) of deoxyribonucleotide triphosphates (dATP, dCTP, dTTP, dGTP) and 2.5 units of the enzyme, Taq polymerase (obtained from a thermophilic Bacterium Thermus aquaticus-Taq) is added to it. Two primers of about 20 to 30 bases long, complementary to the 3′ ends of the two chains of the template are also added to about 0.25 M of each primer. A few drops of mineral oil are added to seal the reaction and condensation prevention. This total reaction mixture is made to a volume of 100 µl.
The P.C.R is carried out with the above mixture in the DNA thermal cycler which cycles between three temperatures at regular time intervals, automatically. The first temperature in the cycle is maintained at 94°C for 20 sec, which causes separation of the double stranded template DNA strands.
Then the temperature is changed to 55°C for 20 sec, due to which the primers are attached (annealed) with each of the complementary template strands. Then finally the temperature is maintained at 72°C for 30 sec, which is the optimum temperature for the enzyme Taq polymerase that facilitates the enzyme to extend the polynucleotides on the primers and completes the polymerization. This cycling of temperature then continues for 20-30 cycles, thereby causing a polymerase chain reaction and thus producing one million copies in 20 cycles which needs about 40 minutes of time.
The Template:
Template is a double stranded DNA fragment which is to be amplified by PCR. It can be any DNA fragment (or gene) of interest, for instance the beta-globin gene fragment, albumin gene or any one DNA selected for the purpose. This DNA fragment can either be in a pure form or in a homogenous mixture of two or more DNA fragments (i.e. crude form) or may be a part of the complete genomic DNA (chromosome).
It can either be synthetic DNA or native DNA (a natural one). It is very much necessary to know the complete DNA sequence of the DNA fragment that is to be amplified by PCR, because a primer of 10-20 bases has to be added which will be complementary to the 3′ end bases of both the strands and this primer can be prepared only when the base sequence of the DNA template is known.
So upon heating to 94°C both the strands separate and the primers attach to both the ends and the enzyme adds bases, nucleotide by nucleotide complementary to that DNA strand (template) to the free 3′ end of the primer. The minimum number of the template DNA required for P.C.R. is 102 (i.e. = 0.1 µg of human genomic DNA). It can be up to a maximum of 105 DNA molecules (— 2.6 µg). In each cycle of P.C.R., the template doubles exponentially, thereby doubling the rate of P.C.R in each and every cycle.
The Primer:
The primer is a small DNA fragment of 10-20 bases long, synthesized chemically and is complementary to the base sequence of both the template DNA strands at the 3′-ends. To synthesize this primer the sequence of DNA that has to be amplified should be known, so that complementary primers can be synthesized. The concept is that, a particular gene (DNA fragment) will have a specific sequence, so preparation of the primer complementary to that specific sequence results in amplification of that particular DNA (gene) whether it is in a crude mixture or as a part of DNA within the genomic DNA.
When heated to 94°C the two template strands are separated and on cooling to 55°C the primers, being more in concentration and shorter in length will anneal to complementary template strands at 3′ ends. The concentration of primer in the P.C.R ranges between 0.05pM to 0.1 pM of each oligonucleotide primer.
The Enzyme:
The enzyme used in polymerase chain reaction is a special type of DNA polymerase known as Taq polymerase. This enzyme does not denature even at high temperatures of 98 °C and thus is said to be ‘thermo stable enzyme’. This enzyme is found (synthesized) in thermophilic (growing in hot springs) bacteria —Thermus-aquaticus abbreviated as Taq. Its molecular weight is 94 KDa and its optimum temperature is 72°C. It is also active in the temperature range of 22°C to 89°C.
Separation of the two DNA strands is carried out by heating the reaction mixture to 94 °C which does not denature the enzyme Taq polymerase, whereas its activity is retained and upon cooling, the enzyme starts polymerization of nucleotides and when its optimum temperature (72°C) is kept, the enzyme would have acted totally for about 3 minutes and by this time it will add 200 to 20000 Kbp.
Hence the time given for each step is very short i.e. sufficient for complete polymerization. This enzyme needs magnesium ions (2.0 mM), dNTPs (0.7 to 2.4 mM), KCl (50 mM) and an optimum pH of 8.4 for its activity. Higher concentration of magnesium, dNTPs (4.6 mM) and KCl (200 µM) will inhibit the activity of Taq polymerase. The conventional protein denaturing agents like urea, ethanol, form amide, at lower concentrations do not have any effect of denaturation on Taq polymerase.
The DNA Thermal Cycler:
The polymerase chain reaction needs three different temperatures:
(1) 92- 97°C for 20 sec, for denaturation of DNA and strand separation,
(2) 40-60°C for 20 sec, for primer annealing and
(3) 65-80°C for 30 sec, for primer extension or polymerization by Taq polymerase.
The total time per cycle of P.C.R. comes to about 3.75 minutes, which includes the time needed to reach each and every changed temperature and the time interval for each of the temperatures. In this short time interval three different temperatures have to be changed and maintained for a specified time.
Therefore an automatic DNA thermal cycler is prepared which heats up water by electric current resistance and then the cooling is effected by fluid flow in the cycler (refrigeration). To this is attached a temperature sensitive knob which automatically switches on/off in a cyclic manner in the specified time limit, for that particular temperature.
Uses of P.C.R.:
(1) It is used to alter a particular template sequence for production of newer and desired DNA, obviously by getting ample number of DNA copies.
(2) A particular DNA sample can be isolated (by amplification) from a crude sample of DNA which has got great research applications, medical diagnostic applications and forensics.
(3) To detect any defect in DNA sequence either hereditary or infected by virus/bacteria. PCR has been used to detect sickle cell anemia, mutation, HIV genomic sequence infection, and altered sperm genetics by DNA amplification.
(4) PCR is an effective procedure for detecting the presence of a known DNA sequence in very small, crude samples, without purification. Due to this, PCR can be used to determine whether a particular illness is due to a viral infection. If the sequence of the viral DNA is known previously, then a pair of primers that anneal to sites in the targeted viral DNA can be synthesized. After PCR cycling a DNA fragment of a specific size will be amplified only if the viral DNA is in the sample, if not no amplification.
(5) PCR is used to detect naturally occurring mutations.
(6) It is also used to produce mutations artificially.
(7) To assemble whole genes from synthetic DNA oligonucleotides, PCR is used.
(8) PCR is also used for DNA sequencing.
(9) PCR is used in DNA finger printing technique.
TB-PCR-RNA:
The bacterium Mycobacterium tuberculosis (TB) has been found only in humans. The disease spreads directly between people through the air. Every second, someone somewhere in the world gets infected, and each year TB kills about 2 million people. The ancestor of contemporary Mycobacterium tuberculosis originated from a 3 million years old species. Mycobacterium tuberculosis bacilli are inhaled through the lungs to the alveoli, where they are phagocytosed by polymorphonuclear leukocytes and macrophages.
Although most bacilli are initially contained, some are carried to the region’s lymph nodes. Eventually, the thoracic duct may deliver mycobacteria to the venous blood; this may result in seeding of different organs, including the kidneys. The genitourinary system is a common site of extra pulmonary tuberculosis (TB).
Genitourinary tuberculosis (GUTB) may involve the kidneys, ureter, bladder or genital organs. Clinical symptoms usually develop 10-15 years after the primary infection. Only about a quarter of patients with GUTB have a known history of TB; about half of these patients have normal chest radiography findings.
PCR is a very sensitive diagnostic tool for TB. As the TB is widespread over the complete human body, sample from any part of the body can be used to conduct PCR. PCR-RNA Enzyme Immunoassay may detect mycobacteria. However, polymerase chain reaction DNA enzyme immunoassay of blood may be a sensitive means of diagnosing mycobacterial infection.
The following PCR tests are available with near-equivalent quality:
i. Genus-specific 16S rRNA PCR test
ii. Species-specific IS6110 PCR test
iii. Roche amplicor MTB PCR test
iv. Amplified Mycobacterium tuberculosis direct detection test (AMDT)
RNA extraction and reverse transcriptase (RT) PCR:
The mRNA extracted from the sample is used as a template for reverse transcription followed by PCR. RNA is extracted from the strain infecting the patient, for instance H37Rv cells, cultured in vitro in Middlebrooks 7H9 medium supplemented with albumin-dextrose complex and dissolved in 50 pi of nuclease-free water. The first-strand synthesis is carried out using avian myeloblastosis virus reverse transcriptase. This is followed by heat denaturation to inactivate the enzyme. Subsequent second-strand synthesis is done using Tfl polymerase.
The upstream primers for DNA of H37Rv are:
The PCR product is visualized by electrophoresis in a 1% agarose gel. A 597-bp RT-PCR product is observed upon staining with ethidium bromide. A 600-bp DNA molecular size marker is run alongside.
Application of Biotechnology # 5. Subunit Vaccines:
Subunit vaccines are the specific cell surface antigens, injected into the body, which produce immunity against this infectious agent and its other related species.
“Prevention is better than cure”—hence medical sciences has coined the term ‘prophylaxis’ which underlines the methods for prevention of diseases from emergence and spread. Vaccination is one among them. Its concept and mechanism is described hereunder.
Immunological System:
All the components present inside the body, specially the proteins and oligosaccharides attached to it, are recognized as ‘self by the internal environment of the body. Any compound entering inside an individual from outside is recognized as ‘foreign body’ by the internal mechanism of the person.
This foreign body can either be a protein, oligosaccharide or any other compound, preferably with a molecular weight more than 5000 kDa. This foreign body is immediately destroyed by the natural biochemical mechanism of the body, which is known as bodies’ defense mechanism or immune system. Failure to modify, destroy and detoxify the foreign body especially those of viruses and bacteria leads to various infections, which may either be hazardous to health or lethal.
Natural Defense Mechanism by the Body:
The foreign substance termed as ‘antigen’ entering into the circulation is first recognized by the plasma Antibody Presenting Cell (APC) named as macrophage, which takes up the antigen by phagocytosis, then hydrolyzes it and either kills it, or processes it by cleaving it at all the points except at a particular sequence, recognizable by other cellular receptors, which is a particular pattern of amino acids (or sugars) called epitope. This epitope is presented on its cell surface, in combination with some proteins called ‘Major Histocompatibility Complex (MHC)’. The antigen engulfed macrophages also produce some protein messengers called interleukins or lymphokines.
Further action on the antigen is carried out by specialized cell called lymphocytes. The lymphocytes present in the blood circulation are of two types (1) B-lymphocytes (B-cells) and (2) T-lymphocytes (T- cells). Once the interleukins and MHC-epitope complex on macrophages are found in the circulation.
The T-cells start responding to it and differentiate themselves into four types:
1. T-helper cells (Th-cells): These cells help in differentiation of B-cells specific to this antigen.
2. T-suppressor cells- (Ts-cells): These are the cells which suppress the activity of B-cells.
3. T-cytotoxic cells (Tc-cells): They kill or lyse the infected cell.
4. T-delayed type hypersensitive cells (Td-cells): These cells are involved in delayed hypersensitive reaction.
The differentiation of a T-cell in response to a particular antigen, into the above mentioned T-cells takes place by DNA rearrangements and the differentiated cells help in clearing that particular antigen/ infection, either directly or indirectly. Once the infection is cleared these specific T-cells degenerate except some cells which are stored as ‘Memory T-cells’ for that particular antigen.
Simultaneously, the antigen is also specifically recognized by the B-lymphocytes by their cell surface receptors called immunoglobulin’s (Ig). These immunoglobulin’s are also secreted into the blood by the B- cells. There are five types of immunoglobulin’s-(a) IgG (b) IgM (c) IgA (d) IgD and (e) IgE.
The IgM receptors on the B-cells, present non-specifically, first recognize any antigen, bind to it and takes it inside the cell by phagocytosis, degrades it into small pieces (or peptides) and presents it on its cell surface in the processed form in combination with major histocompatibility complex-II. This complex is recognized by the Th-cells (produced in response to the same antigen) which induce (through interleukins secretion) the differentiation of the B-cell into a mature antibody secreting plasma cells. This differentiation of B-cell takes place by the rearrangement of the cell DNA by a process called ‘transposition’, which arranges the genes in such a way that the antibody gene product thus produced i.e. the protein immunoglobulin, is complementary to the antigen, which results in antibody binding specifically to the antigen thereby precipitating it, thus resulting in the inactivation of the antigen.
Most of these plasma secreting B-cells degenerate excepting few cells which are stored as ‘Memory B-cells’ for that particular antigen. Thus B-cells inactivate an infection (antigen) and T-cells clear an infection (antigen). Both types of cells are generated in response to an infection (antigen). After the infection is over, both the B and T-cells are stored as memory cells containing a number of Immunoglobulin receptors on B-cells and marker recognition receptors on T-cells, both specific and of high affinity to that particular antigen, as compared to the undifferentiated cells.
When the same infection (antigen) enters the body for the second time, these memory cells come into play and the immune response is elicited at a faster rate and in a shorter time interval, than that of the first instance of infection, because the T and B cells are already differentiated and their DNA rearranged for this specific antigen. This is known as an immune response by the individual to a foreign body or an infection.
All these processes i.e.:
i. Identification of the antigen by the macrophages
ii. Endocytosis of the antigen by the macrophages
iii. Processing of the antigen by the macrophages
iv. Presentation of the antigen epitope-MHC on the surface of macrophage
v. Production of interleukins by macrophages
vi. Interleukin response by T-cells
vii. Epitope-MHC recognition by T-cells
viii. Differentiation of T-cells into Th, T , Ts, Td cells
ix. Killing and clearing of antigen by T-cells
x. Recognition of the antigen by B-cells
xi. Processing of the antigen by B-cells
xii. Presentation of the antigen epitope-MHC-II receptor on B-cell surface
xiii. Recognition of epitope-MHC-II B-cells receptors by Th cells
xiv. Differentiation of B-cells into mature immunoglobulin secreting plasma cells
xv. Inactivation of the antigen by formation of antigen-antibody complex take a considerable duration of time interval (from 2-4 days).
If the rate of multiplication of the infection (virus or bacteria)/antigen is faster than the rate of growth of the immune response, then it results in the development of the disease. On the other hand if the immune response is faster than the multiplication of the virus or bacteria, then it results in the prevention of the disease by the immune/natural biochemical mechanism of the body.
In the case of the antigen (virus or bacteria) entering the body for the first time, the time required is very high to elicit any immune response. So depending upon the infected antigen, i.e. whether it is fast growing or slow growing, the person either becomes resistant to it or succumbs to its disease condition. The disease may be hazardous to heath or lethal.
In the case of a second infection by the same virus/bacteria/antigen, the immune response is much faster than the first infection, due to the presence of memory cells specific for this antigen. These memory cells can make the person immune within few hours or in a day or two, depending upon the infective agent, thereby preventing the emergence of the disease.
Among the various viruses/bacteria infecting the human population, some are very wild and vigorously multiplying in the body, such that the immune response becomes incapable of controlling it, leading to ill health, permanent disability or loss of life. On the other hand, some others are slow growing and are easily controlled by the immune system of the body, hence it essential to vaccinate against fast growing microbes.
Modern Vaccines or Subunit Vaccines:
Sub-unit vaccines are specific cell (viral or bacterial) surface antigens, when injected into a person, will produce immunity against this agent and other related agents. Sub-unit vaccines are made from one or a few components of the organism, instead of the whole organism.
The modern vaccines or subunit vaccines are safe, efficient and cost effective, which are a result of the scientific knowledge relating to the structure of the genomes of many organisms, structure and functions of antigens for the causative viral disease, rDNA technology, gene cloning and the selection of antigens by the expression of the gene product.
Human diseases for which biotechnology offers new or improved vaccines are:
i. For viral diseases like measles, mumps, rubella (MMR), polio, tuberculosis, hepatitis, pneumonia, influenza, rabies.
ii. For bacterial diseases like enteritis, neonatal scours and clostridialtoxicoses.
iii. For parasitic diseases like malaria, leishmaniasis.
Design of Various Modern Subunit Vaccines:
Sub unit vaccines are produced by selecting a particular viral capsid protein/polysaccharide which is antigenic.
The immunogenic components for a few of the disease causing organisms are:
i. HBsAg for hepatitis B
ii. Capsid protein VP3 for hepatitis A
iii. Pneumococcal capsular polysaccharide for pneumonia
iv. Haemagglutinin and neuraminidase for influenza
Methods:
The following methods are adopted for the production of a subunit vaccine:
1. Protein extraction from whole virus culture:
The virus is cultured in in-vitro culture media in large amounts and the protein is extracted from it and is injected into the person to elicit an immunological response and thus generate memory cells to whole virus, which neutralize the viral infection.
2. Use of rDNA technology for production of capsid protein:
The viral genomic RNA is transcribed into DNA, cleaved by a restriction endonuclease, inserted into a plasmid vector, which is also cleaved by the same restriction endonuclease, so as to produce the rDNA and is cloned in yeast cell as the host. Cloning of the rDNA in yeast cell produces glycosylated structural proteins but not in E. coli, which gives un-glycosylated proteins. Thus large amounts of immunogenic proteins are obtained by gene cloning, which is injected to give protection to the human beings against the disease.
The pure individual protein injected, though succeeded in eliciting an immune response in the vaccinated individual, it was much less immunogenic than the intact whole particle, because the pure protein created B-memory cells but not the T-cells.
In order to enhance the immunogenic activity of the separated proteins, a mixture of plant glycoside saponin, cholesterol, and phosphatidyl choline is added to many copies of the protein to form a cage-like structure, mimicking the natural microorganism. These complexes known as immuno-stimulating complexes have antigenic activity equivalent to the viral vector.
There is further development in the preparation of sub-unit vaccine for virus. In the virus capsid protein only few amino acids are truly antigenic/immunogenic present at few regions in the protein and these regions are termed as ‘epitope’. Therefore synthetic peptide is made with these amino acids in the same sequence as that of the epitope. Eight such peptides when linked together by lysine residues to form a frame work (immuno-stimulating complex) will be equally good as an immunogen as that of the whole viral particle itself.
Further a synthetic DNA can be prepared which can code for these amino acids and joined to the gene coding for hepatitis B core protein. This rDNA produced a chimeric protein which takes a spherical shape of about 22 nM in diameter. This protein, obtained by cloning in Autograph californica as the host, acted as a sub-unit vaccine and protected at par with the whole viral particle vaccination.
3. Recombinant virus as a sub-unit vaccine for different diseases:
Vaccinia virus exists in two forms or in other words it has got two different strains, one of them is infectious and hence is termed as virulent form of vaccinia virus and the other one is non-infectious, which is termed as non-virulent vaccinia virus.
The non-virulent form is extensively used as a vehicle or agent to introduce recombinant antigenic epitopes for different diseases. The other agents used are 17D strain of yellow fever virus, Salmonella typhi murium, Bacillus Calmetta Guerin (BCG) and poliovirus. All these are attenuated vaccine vectors used as delivery vehicles for various sub-unit vaccines.
Viruses causing a particular disease will exist in a number of different strains. Therefore vaccination for a disease by one strain results in invasion by the virus of another strain causing the same disease. Therefore an effective sub-unit vaccine can be prepared by rDNA technology wherein antigenic epitopes from all the strains are mixed with a plasmid vector and joined into a single recombinant DNA and then transferred into any of the agents mentioned above—for instance a virulent vaccinia virus, which will produce the antigenic epitopes for all the strains of a particular disease causing virus or bacteria, on its capsid coat. This is known as recombinant vaccine.
Therefore, vaccination with this recombinant vaccine agent (vaccinia virus with various antigenic epitopes) results in development of immunity to a disease (or many diseases) for all the strains causing that disease. Non-virulent vaccinia virus can also be used as an agent to introduce epitopes for two or more disease causing viruses with epitopes for all of the strains. Such recombinant vaccines are prepared for Rabies.
4. Other sub-unit vaccines:
Virulence genes of enteric pathogens have been excised or modified and this recombinant organism colonizes in the intestinal mucosa and will express surface antigens which prevent adherence of natural virulent bacteria thus preventing the disease.
A 68 kDa outer membrane protein is present in virulent strain of bordetella bronchi stptica, but not in avirulent strain, which is the main immunogenic site. This protein is cloned in the avirulent strain or some other host, thus producing a cost effective subunit vaccine.
For any disease causing antigen—the epitope is chemically synthesized as a peptide and injected into the body. The antibodies produced to this are Ab1, which are used to produce anti-Ab1 antibodies i.e. Ab2 or anti-idiotypes. Ab2 will be structurally similar to the epitope of the original antigen. Therefore this anti-idiotype Ab2 is produced in large quantities and used as sub-unit vaccine against that particular disease. Vaccines are more commonly produced by this method, as monoclonal antibodies by hybridoma technique.
5. DNA vaccines:
The most recent approach in the vaccine design is the use of synthetic DNA for the epitope of an antigen and produce a recombinant DNA, insert it into a non-virulent viral vector and inject it into the person. This virus proliferates in the body expressing the epitope of interest, thereby producing immunity to the disease for which the epitope is designed. This reduces the cost and labour of producing the antigenic protein/epitope peptide, its extraction, purification and characterization.
Subunit Hepatitis B Vaccine for Human Use:
The surface antigen of hepatitis B virus (HBsAg) is a protein consisting of 226 amino acids with a mol. weight of 25,398. A synthetic nucleotide sequence of 892- base pair is prepared and inserted into a plasmid vector. The portion of the gene coding for this protein does not contain any intervening sequences. This recombinant DNA is introduced into the yeast Saccharomyces cerevisiae.
The antigen is harvested and purified from fermentation cultures of a recombinant strain of the yeast containing the gene for the adw subtype of HBsAg. The fermentation process involves growth of Saccharomyces cerevisiae on a complex fermentation medium which consists of an extract of yeast, soy peptone, dextrose, amino acids and mineral salts.
The HBsAg protein is released from the yeast cells by cell disruption and purified by a series of physical and chemical methods. The purified protein is treated in phosphate buffer with formaldehyde and then co-precipitated with alum (potassium aluminum sulfate) to form bulk vaccine adjuvant with amorphous aluminum hydroxyphosphate sulfate. Hepatitis B vaccine (Recombinant) is a non-infectious subunit viral vaccine derived from hepatitis B surface antigen (HBsAg) produced in yeast cells.
Application of Biotechnology # 6. Organ, Tissue, Cell Culture:
Growing organs, tissue or cells in laboratory dishes, outside the body is known as cell, tissue and organ culture. If individual cells (bacteria, viral or separated animal/plant cell) are grown in culture media then it is known as cell culture. If a group of cells in a tissue are grown in laboratory glass then it is known as tissue culture. If the whole organ is cultured in culture media/laboratory glass then it is known as organ culture.
Every cell present in the human body is not capable of growing in laboratory, only a few types of cells can grow in vitro. Cells that can be grown in culture are tumour cells, steroid producing adrenal cells, ACTH-secreting pituitary cells, insulin secreting pancreatic islet cells, growth hormone and prolactin secreting cells from pituitary tumour, melanocytes, neural cells, epithelial tissues, skeletal tissue etc.
Those cells which can be grown in vitro are neither suitable for industrial use nor for scientific purpose because many cells die during the course of time releasing toxic substances which inhibit the activity of other live cells. Hence, in order to avoid this problem and to achieve an exponential cell growth, the cells are converted into immortal cells called ‘cell line’. The following is the procedure for the production of cell lines.
Procedure for Production of a Cell Line:
A piece of tissue is removed from an organism. Then the adhesion between cells is broken with enzymes like trypsin or collagenase. These cells are then transferred to a plastic dish or bottle. The dish/bottle contains nutrient solution, known as culture media (made up of appropriate salts and nutrients). Then the cells are incubated at 37 °C in 5-10% CO2 to aid in pH maintenance and perfused with 90-95% oxygen.
Human or animal cells grow only when they actually touch a solid support, they are not free floating. The solid supports generally used are plastic, glass, Teflon, DEAE-Sephadex, etc. all of which are transparent and aid in microscopic observation. Further any manipulation relating to the cells or tissue is done in a laminar flow hood, so as to prevent any contamination with microorganisms.
When provided with all the suitable requirements the cells grow, divide and cover the surface of the container and look like the tissue from which they are derived. This culture is referred to as primary cell culture, because all cells touching other cells or the walls of their container will stop dividing due to contact inhibition (which can be overcome by growing cultures on micro-carriers like anion exchange resins).
If this primary culture is diluted two fold and the culture transferred to fresh medium, the cells will again start growing. This type of repetitive culturing of the cells is limited because the growth of animal cells ceases after about 50 cell divisions, either due to lack of proper culture media or built-in-senescence mechanism.
However, some among these cells in each culture continue to grow after numerous transformations, but these cells are not normal as they undergo some chromosomal abnormalities and these are termed as diploid cell strains and are not different from cells of the primary culture. After sometime, even the diploid cell strains lose the ability to grow, but once again a few cells among them will survive and are termed as heteroploid cells, because they undergo many chromosomal rearrangements and deletions.
These cells will grow in culture indefinitely as long as the medium is replenished, becoming effectively immortal. These survivors are known as ‘cell line’. They are said to have undergone transformation. Transformed cell lines grow indefinitely, exhibit heterogeneity, lose contact inhibition and do not require solid support for growth. A tumour tissue represents a transformed cell line. The most famous and the oldest cell line is the Hela cell line, derived from human cervical cancer cells, which are growing since decades and creating problems in tissue culture laboratory.
Purpose of Cell and Tissue Culture:
(1) Cultured cells are used as substitute hosts to study the pattern of viral infection.
(2) Cell lines are used in the manufacture of vaccines, antibodies, hormones, interferon’s, urokinase enzyme, vitamins, steroids etc. on a large scale industrial basis.
(3) They are good tools for testing the potency of drugs.
(4) These cells also serve as models to study the metabolism of various substances.
Culture Media:
Culture media is the environment provided for the growth of the cells in laboratory, similar to those conditions that the cell has been exposed to in vivo (i.e. inside the animal body). When a cell is removed from its original tissue or organism and placed in the laboratory glass for culture (i.e. multiplication) it will not proliferate, differentiate and divide until and unless it is provided with all those components and substances upon which it was surviving in the tissues or organism.
These components and substances, required for the proper growth and maintenance of the culture is known as culture media. The culture media should contain a support or matrix (termed as physical media) and appropriate nutrients, hormones and stromal factors (the chemical media), for survival and growth of the cells in vitro.
Serum is the most economical, easily available and most widely used culture media for animal cell culture. It provides all the necessary nutrients and substances to sustain a culture. Serum is an extremely complex mixture of many small and large biomolecules with different, physiologically balanced growth promoting and growth inhibiting activities. Fetal calf serum is the preferred serum as culture media.
The major serum components are—glucose, urea nitrogen, proteins including albumin, macroglobulin, fibronectin, uric acid, creatinine, haemoglobin, bilirubin, alkaline phosphatase, LDH, insulin, TSH, FSH, growth hormone, prolactin, T3, cholesterol, cortisone, testosterone, progesterone, PGE & F, vitamin A & E, Na, K, CI, Fe, Zn, Cu, Mn, Co, V, Mg, Se, Ca, P, etc.
The major functions of serum as a culture media are—to provide nutrients, hormones, growth factors, attachment and spreading factors, binding proteins, vitamins, minerals, lipids, protease inhibitors and pH buffer. Though serum is widely used culture media, it has some disadvantages like all cells in the animal body do not come in direct contact with blood, some enzymes present in serum can convert the cell secretions into toxic compounds.
Serum collected in different batches differs. The constituents present in it may be inadequate for maintenance of the culture and virus, fungi, and bacteria may contaminate the media easily. Hence, recently culture media are developed artificially by mixing appropriate nutrients and chemicals, so as to provide an equivalent environment for culture.
There are three types of artificial culture media:
(a) Serum-free culture media: i.e. No serum but some proteins extracted from serum are supplemented.
(b) Protein-free culture media: i.e. All the constituents of serum, without serum proteins.
(c) Chemically defined media: It contains substances of small molecules and genetically engineered proteins or peptides.
The various artificial culture media available commercially are Eagle’s minimal essential medium (MEM), Dulbecco’s modified enriched medium (DME), Ham’s F-12, CMRL 1066, RPMI-1640, McCoy’s 5A, IMDM and MCDB-301.
The main components of the chemically prepared artificial culture media are-minerals and trace elements like Cu, Zn, Co, Mn, V, Se, Mg, Al, Ba, Cr, Ge, Ti, Sn, Ni etc, vitamins-C, E, and B-complex. Carbohydrates as energy source like glucose, or pyruvate as a source of acetyl-coA. Lipids like cholesterol, long-chain fatty acids, glycerides and lipoproteins. Amino acid, proteins like albumin, transferrin and synthetic polymers, hormones and growth factors plus extracellular matrix.
Krebs’s ringer bicarbonate solution is also used as a culture media which is Composed of NaCl, KCl, MgSO4, CaCl2, NaHCO3, KH2PO4, gassed with O2-CO2 mixture. It also contains balanced salt solutions, vitamins, carbon source like mannitol, amino acids, plasma proteins and antibiotics.
Cell Sorting:
Cell sorting is the process of separating the cells of interest from the unwanted cells. Cell sorting is done at two levels in developing a cell line, (1) While selecting a cell from a tissue and (2) While shifting the culture from primary cell culture to secondary cell culture (diploid cell) to tertiary cell culture (heteraploid cell) to the immortal cell line (transformed cell).
Cells in a culture die continuously beginning with the primary culture to the diploid culture to heteraploid culture to the immortal cell lines. However some cells remain live in each of the stages of the cell transformation. Hence cell sorting is a process of separating the live cell from the dead cells in each of the culture types. The procedure adopted for cell sorting in the culture medium are—chemicals which can dissolve dead cells easily are used wherein the dead cells dissolve leaving the live cells intact.
Other methods used for cell sorting are density gradient centrifugation, electrophoresis, isoelectric focusing sorting or separation of cells can also be carried out using magnetism, complement lysis of specific antibody tagged to unwanted cells, flow cytometry (one of the best methods of cell sorting) and affinity chromatography.
Cytofluorimetry is the most recent and widely used cell sorting technique. In this the cells are stained with a fluorescent dye, wherein the dead cells get stained and the live cells are unstained. Depending upon the fluorescence, the cells are separated and hence the method is termed as fluorescence activated cell sorting (FACS).
Cell Counting:
The counting of the viable (live) cells in a culture is done to determine the number of viable cells/ml of the cell suspension, so that the exact and optimum dilution of cells may be made, which would be suitable for the cell growth. The viable cell count is achieved by staining the living cells by using a vital dye like neutral red (NR). Trypan blue may also be used, which stains only the dead cells. The stained cells are counted by using Neuberger counting chamber. Cell counting is also done by electronic particle counter and Coulter particle counter.
Cryopreservation of Cell Lines:
When dealing with established cell lines, it is essential that a set of cell line be preserved so that if the culture becomes unsuitable for use due to contamination, loss of growth, defects in incubation etc. resulting in loss of the culture, then the preserved cells can be used and thus the cell line is not lost. In addition, freshly trypsinized/collagenized cells, cells of secondary culture, embryonic cells, human placenta or fetus and other such cells which are difficult to procure at will are preserved so that they may be used afterwards. Cells are preserved by two methods:
Freezing with Glycerol:
The cells at a concentration of 2 x 106/ml are taken in Eagle’s medium containing 20% calf serum and 5% glycerol and distributed in aliquots of 2-4 ml in 5 ml vials, which are then frozen so that there is a slow fall in temperature of 10° C/minute till —25° C. The vials are then placed in liquid nitrogen and stored at -196° C.
Freezing with dimethyl sulfoxide:
2 x 106 cells/ml are taken in Eagle’s medium containing 15% calf serum and 10% dimethyl sulfoxide and distributed in 3-5 ml vials and stored in liquid nitrogen at – 196°C or at -70°C.
Application of Biotechnology # 7. Hybridoma Technique:
Hybridoma is an immortal cell or cell line, formed by the fusion of a myeloma cell with an antibody secreting plasma cell (lymphocyte). Hybridoma technique is the process of producing a hybridoma cell. Antibody raised to a particular antigen in the serum is used for many purposes like the detection of presence of a toxic substance in clinical samples and also for its quantization, localization and purification.
Any antigen will contain two or more epitopes or antigenic parts and the immune system differentiates in such a way that many plasma cells (B-cells) are differentiated specific to each epitope and each B-cell will produce an antibody having affinity to only one epitope on the antigen.
Therefore different B-cells will produce different antibodies specific for the different epitopes on the same antigen. These many different antibodies produced by different B-cells for a single antigen are termed as polyclonal antibodies as they are produced by clones of different cells.
The use of polyclonal antibodies as diagnostic and therapeutic agents was not specific because antibodies produced to the same antigen differ from batch—to—batch due to the difference in the response of the immune system in the body. Therefore Monoclonal Antibodies (MAb) were produced i.e. those antibodies which are identical and specific to a single epitope of the antigen and produced by a single clone of B-cells.
B-lymphocytes producing a specific antibody (MAb) are incapable of growing in culture. Hence a hybrid cell type was created having the B-cell genetic components for producing this specific antibody and the cell division functions of a similar type of cell so as to enable the hybrid cells to grow in culture.
Some B-lymphocytes, naturally become cancerous (myeloma) and hence become able to grow in culture. So these myeloma cells were used to fuse with the antibody producing B-cell forming the hybridoma cell capable of growing in culture indefinitely and secreting the specific MAb.
The Technique:
When cells from two different sources are mixed and incubated with certain surface- active agents like polyethylene glycol (PEG), sendai virus or high voltage direct current pulses, then the cells fuse to form heterokaryones i.e. a single cell having two nucleus from different cells.
The nuclei of some heterokaryones fuse to form hybrids. If one of the cell forming the hybrid is immortal (i.e. capable of growing indefinitely in culture) and the other is mortal (i.e. incapable of growing in culture), then the hybrid will be immortal and can be maintained indefinitely.
Production of Monoclonal Antibody (MAb):
Monoclonal antibody (MAb) producing immortal hybrid cells (hybridomas) are produced by injecting mice with a particular antigen and an immune response to this antigen is developed in the animal. Then the mouse is killed, spleen is taken and the cells are separated.
Only the spleen B-lymphocytes (as source of antibody) of this immunized mouse are mixed with cultured mouse myeloma cells (tumour cells) and incubated with 35% polyethylene glycol for few minutes. This procedure will not fuse all the cells, instead only a few cells are fused. So the incubated mixture is expected to contain myeloma cells, spleen cells, myeloma-myeloma fusion cells, spleen-spleen fusion cells and a few of them will be hybridoma cells i.e. myeloma-spleen fusion cells.
In order to isolate the fused cells (hybridoma cells) producing antibody of interest, first of all the hybridomas are detected and separated from the other un-fused cells. The spleen B-cells, however die as they cannot grow in culture, whereas the myeloma cells are selected and separated by growing the cell mixture in a medium containing Hypoxanthine (H), Aminopterin (A), and Thymidine (T), hence known as HAT medium.
Myeloma cells are devoid (mutant) of the enzyme-Hypoxanthine Guanine Phosphoribosyl Transferase (HGPRT–), so they cannot naturally utilize bases by scavenging process and the de-novo synthesis of bases is inhibited by aminopterin (in the HAT medium), hence the myeloma cells die, whereas the hybridomas survive because they have the enzyme HGPRT , coming from the B-lymphocytes. Therefore growing cells in HAT medium for 10 to 14 days after fusion results in spleen B-lymphocytes-myeloma hybrid cells to survive and all others die.
Among these hybrid cells, the cells producing antibody against the immunizing antigen are identified and separated by Enzyme Linked Immunosorbant Assay (ELISA) or solid phase radio-labelling. For this, dilution culture is taken up such that each cell is grown in a different culture to form a clone of single type, the secretions of these cells present in the culture media is taken as sample for ELISA or solid phase radio-labelling and the hybridoma cells are selected.
After screening and selecting the hybridoma cells producing the antibody of interest, they are cultured by soft agar cloning or dilution cloning, which secrete monoclonal antibodies (MAb) highly specific for the epitope of the injected antigen.
Culturing hybridoma cells producing MAb in laboratory culture dishes produces less number of antibodies, hence in order to get large quantities of antibodies in short time, the hybridoma cell line is grown as intra-peritoneal ascitic tumour, induced by mineral oil injection (pristane) into mice. Once cloned in the ascitic tumour, all growing cells will produce antibodies in large amounts (i.e. 1-2 mg/ml).
Uses of Monoclonal Antibodies:
(1) Diagnosis or treatment of infectious diseases, tissue typing for organ transplantation and blood grouping.
(2) Calculation and differentiation of human lymphocytes at various stages of development.
(3) Analysing mixtures of complex antigens.
(4) Determination of the structure of cell membranes and membrane proteins.
(5) Labelling and identification of specialized cells.
(6) Tumour and cancer detection and therapy.
(7) Understanding the mechanism of antibody diversity generation at DNA level.
(8) Monoclonal antibody based assay is replacing the immuno-florescence assays.
(9) MAb are used as tools in enzyme purification and genetics.
(10) Used to isolate receptors, non-histone chromosomal proteins, hormones etc.
(11) Used to treat diseases like tetanus, snake bite, rabies, herpes-B, leukemia virus etc.