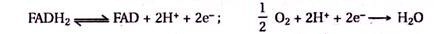This article provides an overview on plant diversity and its monitoring through DNA amplification.
Introduction:
Perhaps, the most intriguing part on earth is its immense biological diversity. In twenty-first century, why one should be concerned about it?
The Answer Lies in its Three Main Components:
(i) Genetic (DNA) diversity,
(ii) Species diversity and
(iii) Ecosystem diversity.
When one discusses biodiversity as a whole these three components are tackled together. Genetic diversity is the first step in the process where a base mutation, in a suitable locus, could lead to a new species. Continuous inbreeding often unbalances the genetic make-up of a species by promoting admixture of genes.
Accumulated knowledge by population genetics indicates that each species has its own inherent gene diversity (Houerou, 1997) that diverges through natural selection. The species diversity and ecosystem diversity then follow. In ecosystem, even soil microbes can determine diversity within plants and animals—by their interactions with them (Huston, 1979; Dobson and Crawley, 1994; Hooper and Vitousek, 1997; van der Heijden et al. 1998).
So, biodiversity is the total gene pool or genetic polymorphism in an area, and ecosystems have the collection of all organisms within a particular area each differing in physical structure, genome composition and gene function (Tilman and Downing, 1994).
Along a latitudinal gradient, species diversity tends to increase toward tropical areas. Within tropical areas, species diversity increases along a longitudinal gradient. Availability of nutrients is also another main factor for species diversity (Norse, 1993). The picture is the same on land, rivers, and delta. Therein sunlight, nutrients, and biotic interferences determine diversity. It ultimately leads to evolution.
Land and aquatic ecosystems are highly dynamic—though they are often disturbed by biotic interferences. Coral reef though has an entirely different ecosystem, it is also subjected to outside interferences. On the other hand, deep-sea ecosystem has less outside interferences.
It may be noted that a minimum amount of genetic diversity within a population is essential for a species’ survival (Boyce and Catena, 1991). Healthy ecosystem supports high biological diversity, on the other hand, stressed, unhealthy, or highly disturbed ecosystems do not.
By recent DNA techniques these can be monitored (Karp et al., 1998; Harris, 1999). In this article I have discussed the application of various DNA techniques to monitor such diversity. However, a general account of biodiversity is given at the beginning.
Biodiversity in India:
India belongs to pan-tropical region in Asia. Ecosystem diversity in India includes desert, forests, grasslands, and mangroves. Bounded by the oceans at the west, east and south, and the mighty Himalayas on the north, India is one of the 12 mega-diversity regions in the globe and endemism predominates here. Therefore, India possesses rich genetic diversity. For example, out of 20,000 fish species known to the world, 2,200 species of fin fishes are found in Indian waters. Similar is the situation with insects.
Regarding plant population, this country is considered to be the homeland of 167 important cultivated plant species and 320 species of their wild relatives. Major centres for their distribution are delta and grassland areas, the Himalayas (Eastern, North-Eastern, Western), Tamil Nadu hills, and the Eastern and Western Ghats hills. India is considered to be the centre of origin of cultivated rice and possesses 50,000 varieties of rice (about a dozen wild relatives), millets (51 wild relatives), pulses and vegetables (55 wild relatives), spices (27 wild relatives), fiber crops (20 wild relatives), coconut, fruits (104 wild relative), bamboo (more than one hundred species), indigo, taro, tea, coffee and sugarcane (12 wild relatives), etc. (Kumar and Asija, 2000).
Active germplasm holding, at the National Bureau of Plant Genetic Resources (NBPGR, New Delhi), has 1,59,000 plant species. Not much information on algal flora is available from Indian river-lagoon. A recent survey estimates Trachaeophyta (50.9%), Chlorophyta (15.6%), Rhodophyta (25.6%), and Phaeophyta (7.9%), and many of them are being catalogued every month.
Consequences of Threatened Ecosystem:
Since 1980s, people began working to preserve the biodiversity in the globe. The climax had reached in a major UN Conference (UNCED) in June 1992, in Rio de Janeiro, Brazil, where 178 countries participated. That meeting had a major impact to know and preserve flora and fauna of a country. Since then, biodiversity became the slogan for conservation of germplasm (Kumar and Asija, 2000).
The factors which threat biodiversity would change the environment and humans would be in danger, because humans (a small segment of earth’s germplasm) exploit the majority of Earth’s, resources. At present, scientists, media, public and governmental agencies worldwide have begun to recognize the danger from large-scale human interferences that lead to species extinctions.
The rate of this extinction on this globe—particularly in developing and underdeveloped countries—is occurring on an enormous scale—at a rate that had rivalled or even surpassed those of the Cretaceous period when many species including the dinosaurs became extinct. Pimm et al. (1995) estimated that recent extinction rates of species are 100 to 1,000 times their pre-human levels.
At present, every nation is conscious about its natural resources and is trying to catalogue local flora and fauna, especially endemic species by looking at their DNA profiles because unlike other characters DNA is less susceptible to environment and biotic factors.
However, the change at species level first comes at DNA level of an individual. That change is manifested in the phenotypic appearance and adaptability of that individual within a population. Better forms diverge quickly. Even within highly diverse ecosystems, species elements can differ widely — bringing the incompatibility barrier between two or more populations..
Sustainable Use of Plant Diversity:
At present, it is of paramount importance to note the threats to plant biodiversity and to device methodology to counteract them, because plants provide medicine, food, and materials for the industries. The situation will remain unchanged, as long Homo sapiens would survive. If the species richness vanishes from this globe, the very survival of Homo sapiens would be problematical. In order to know and preserve plant diversity, the Botanical Survey of India (B.S.I.) had published Red Data Books of endangered Indian plant species.
Estimation on total number of plant species in this globe varies. Roughly, more than 300,000 angiosperms are catalogued. New species are catalogued in almost every day- especially from neo-tropical and inaccessible regions. Percentage of forest area lost in 14 Asian countries ranges from 34.96%.
In the last century, India has lost 78% of its forests (World Resource Institute, 1990, 1992). Moreover, maintenance of the biological diversity of marine and estuarine systems was largely overlooked, even though it is generally accepted that marine systems are far more species rich and have greater ecosystem diversity than terrestrial systems.
So, maintenance or sustainability of plant resources on land, ponds, lakes, river, and oceans is essential for India’s point of view. Sustainable use of plant biodiversity (of course entire biodiversity) is the need of today’s world because bio-prospecting is the new terminology for the use of microbes, wild flora and fauna.
The Future of Man would depend on Bio Prospecting:
The practical application of biodiversity is bio prospecting, i.e., the search for economically valuable compounds, important genes, or gene products from wild organisms. These compounds have been touted as a source of global economy since the dawn of human civilization. Tropical forests provide raw materials used in industries, help in water conservation and affect the world climate.
Out of 5 to 15 million expected species, only 1.5 million were catalogued, and man has exploited a negligible section of them. Every year newer products are being discovered from already identified species. On this globe, bio prospecting is a never- ending process.
A large proportion of tribal and rural population still depends on wild animals and plants for their basic needs. Also, both modern allopathic as well as traditional systems of Ayurveda and Unani medicines are heavily dependent on plant-based drugs. Unfortunately, extensive exploitation of medicinal plants is offsetting the pharmaceutical industry.
Similar situation would be noted in every plant-based industry. At present, bio-piracy is another aspect that has to be tackled by tropical countries including India. Therefore identification of a species/species diversity and bio prospecting by a quick method such as DNA typing are desirable.
Monitoring DNA-Diversity:
The best way to measure the degree of genetic diversity and measuring genome polymorphism, within a population, species, genus, or higher-level taxon, is a resolution by molecular markers, particularly the DNA and protein markers (Helentjaris, 1989; Caetano et al. 1997; Harris, 1999; Hollingsworth, 1999).
The advantage of a molecular marker over morphological markers is its superior quality, and environment has no effect on these markers during the growth and differentiation of an organism.
The Important Properties of a Good Marker are:
(a) Easy recognition of all phenotypes (homo-or heterozygotes),
(b) Early expression during plant development,
(c) No effect on alternate alleles on plant morphology,
(d) No or low interaction among markers, etc.
Unlike morphological markers, genetic (molecular) markers can fulfill these criteria because rate of evolution could be measured by looking into genetic molecules of related or unrelated taxa. Amongst the two types of molecular markers (DNA and proteins), DNA is superior over protein markers as they are least affected by the environmental fluctuations. Gene sequences are useful to develop molecular markers (Roy, 2003 and Bandopadhyay, 2003).
Often arbitrary sequences are also used successfully to measure genetic diversity (Caetano et al. 1991, 1997). Works of many scientists, who are currently using DNA markers in plant genetics and plant evolution, was timely from the standpoint that this is a rapidly developing technology that can be compared with the nuclear science in mid-twentieth century.
Early Techniques:
Since the dawn of molecular biology, in the 1960s, DNA sequence diversity was measured by re-association kinetics, thermal melting profiles, Southern hybridisation, and Northern hybridisation. Now comes one basic question “Which DNA Marker for what purpose?” As one moves from small to large plant genomes, essentially most DNA “species” can be attributed to repetitive DNA sequences.
Since early 1960s, a eukaryote genome is considered to be composed of different DNA species—unique DNA sequences and repetitive DNA’s (Britten and Khone, 1968). Within the former, message for genes reside. As gene conservation is a rule rather than an exception (gene synteny is prominent in cereals and perhaps in all plants), there would be little variation amongst gene sequences, within related taxa. However, evolutionary rate of repetitive DNA species is many times more than that of a unique DNA sequences.
Moreover, repetitive DNAs often may be highly mobile (transposon). So, often they control gene function (Chaudhuri and Chaudhuri, 1994). By doing so, repetitive DNAs provide tools to Mother Nature to play—to evolve different life forms. So, for an attempt to measure biological diversity, the best bet would be to look into gene-control elements’ sequence-diversity.
The absolute amount of single copy appears to remain meagre in large plant genome where genome replication is the rule rather than an exception. In maize, broad bean (Vicia faba), lily, and in many other plants, no more than a very small percentage of genome appears to consist of single-copy DNA sequences.
It is important to note that many kinetic measurements, with single-copy DNAs, are overestimates because the re-association kinetics was performed at criteria where extensively diverged “fossil” repeats displayed single-copy kinetics. An additional complication is that extensive short-repeat sequence interspersion makes it difficult to find single-copy sequences much larger than several kbp (kilo basepairs) in all but small genomes e.g. Arabidopsis.
At present, any genome structure can be investigated with DNA markers. Restriction fragment length polymorphism (RFLP, Botstein et al. 1980) analysis is the first technique widely used to detect variation even at the gene sequence level. It is a DNA-DNA hybridization technique where a labelled DNA probe is used to identify the level of base sequence diversity by hybridising that probe with template DNA strand.
Another aspect of RFLP is the use of restriction endonucleases that could detect sequence diversity in allied genomes. A probe (marker)-enzyme combination is used to resolute the differences between individual genomes.
The application of RFLP as molecular markers has proven to be a powerful tool for studies in both basic and applied plant genetics and also to study genome evolution. The principal difficulty with RFLP is its reliance on cloning (to produce marker), Southern blotting and Southern hybridization.
For this, one must aim at the development, optimisation and validation of methodologies with special emphasis on high-throughput procedures right from the beginning (e.g. homogenisation of material and DNA extraction). Methods thus developed are to use the probes (markers) to detect the presence or absence of a gene, pedigree analysis, expression of a particular trait, etc. Therefore it is not only time-consuming but costly too. Helentjaris (1989) estimated that one technician could analyse 1,500 genotypes with 40 to 50 markers in 1 year.
PCR-Based Techniques:
Several other techniques for identifying genetic polymorphisms have been described. In 1985, Dr. Kary Mullis and his colleagues developed a unique DNA replication protocol that is known as polymerase chain reaction (PCR) (Mullis and Falcona, 1989). This protocol can be recognized as ‘Photocopying of a DNA molecule’ by repeated DNA polymerisation reactions. It could replace the requirement of cloning for multiplying DNA probes (DNA fragments, marker). In recent years, use of PCR-based markers could solve some of the limitations of earlier RFLP protocols.
Essentially, PCR is an uninterrupted DNA replication protocol where DNA replication is performed by the Taq DNA polymerase; and, instead of a RNA primer, one or two DNA primers (oligonucleotides) are used for this in vitro replication, in an Eppendoff tube. This protocol (now popular as ‘peoples choice reaction’) is so important that the reputed journal Science considered this as the major scientific development of 1989, and had chosen Taq DNA polymerase as the molecule of 1989.
As stated above, the DNA replication process would go for an unlimited fashion, in an ideal temperature (usually 72° C), provided the enzyme, DNA primers (oligos), and four deoxynucleotides (dATP, dTTP, and dCTP) are available. The single strand condition of double stranded reaction products (amplified dsDNA) is achievable by thermal denaturation of dsDNA in a cyclic way.
Therefore, an unlimited supply of amplified DNA products is obtained by PCR. The usual reaction condition is : denaturation at 90 to 98°C (5 to 10 min), two primers that anneal at 34 to 60° C (a few seconds to 1 min), and DNA replication at 94°C (half to 2 min); reaction cycles often vary from 25 to 45— depending on the DNA template.
After denaturation, single stranded DNA primers (oligos) anneal to ssDNA templates, when they are cooled, at specific sites that match their base sequences. If that condition gets enzyme (DNA polymerase) and four essential triphosphates (nucleotides), DNA replication begins, using two ssDNA strands as two complementary templates.
In order to continue the synthesis, the temperature of the reaction mixture is increased and decreased in a cyclic way, with the help of a thermo cycler (PCR machine). Ultimately, from one DNA molecule two dsDNA molecules are synthesized. From two to four, four to eight, and so on. Initially, two DNA template-strands can give two DNA molecules have long chain.
However, subsequently the molecule lengths decrease; the ends become precise when two ends of a synthesized DNA are complementary to two primers. Also, after sixth cycle, the DNA amplification proceeds at an exponential rate. By that way, 2n molecules are produced after ‘n’ cycles. If the process is repeated 20-30 times, in a few hours about billion copies of the sequence flanked by the left and right primers are produced (Fig. 6-1) (Scena and Davis, 1998).
Different PCR Protocols:
In early PCR amplification, use of arbitrary primers RAPD (Random Amplified Polymorphic DNA, Williams et al. 1990) was widespread. It studies DNA loci with the aid of single primers that identify complementary sites on both DNA chains. Unlike the allozyme method, RAPD allows to analyse not only the unique, but also the non-coding DNA portion.
Later, other PCR-based techniques, with single or pair primers, were developed (Bandopadhyay, 2003; Roy, 2003). Among those techniques prominent ones are Arbitrartily Primed Polymerase Chain Reaction (AP-PCR, Welsh and McClelland, 1990), DNA Amplification Fingerprinting (DAF, Caetano et al. 1991), and Cleaved Amplified Polymorphic DNA (CAP) require single primer that recognise specific DNA sequences onto the template ssDNA strands.
In Inverse PCR, amplification of those DNA sequences take place, which are away from the primers and not those which are flanked by the primers. For instance if the border sequences of a DNA segment are not known and those of the vector are known, then the sequence to be amplified may be cloned in the vector and border sequences of vector may be used as primers in such a way that the polymerisation proceeds in inverse direction, i.e., towards the inserted segment, and not away from it towards the DNA sequence of the vector from which primers have been derived.
Anchored PCR uses one specific primer that represent the precise sequences of one of the two ends of the DNA fragment that has to be amplified, because one has the knowledge about the sequence at only one of the two ends of the target DNA sequence. Therefore, anchored PCR utilizes only one primer instead of two primers.
By this technique, only one strand will be copied first : after that a poly G will be attached at the end of the newly synthesized strand. This newly synthesized strand with poly G tail at its 3′ end will then become template for the daughter strand synthesis utilizing an anchor primer which has a poly C sequence linked to it.
Sequence Tagged Sites (STS) and Sequence Tagged Microsatellite Sites (STMSs) can amplify DNA fragments of interest provided the primers are designed on specific DNA sequences (obtainable after DNA sequencing). Primers are designed on flanking sequences of the DNA (or gene) of interest. Therefore, by repeated DNA amplification intervening sequences of a DNA stretch is amplified.
The Multiple Arbitrary Amplicon Profiling (MAAP) was suggested to describe all the characteristics common to these closely related techniques. A more reproducible method, based on the detection of genomic restriction fragments by PCR amplification, is the Amplified Fragment Length Polymorphism (AFLP, Vos et al. 1995), was described. The most favoured one is the Single Nucleotide Polymorphism (SNP).
Presently Popular Markers are SSR, SNP and DNA Microarray or DNA Chips:
Microsatellites, or Simple Sequence Repeats (SSR, Akkaya et al. 1992) have now found popularity as genetic markers for comparative analysis and mapping of genomes. Microsatellite instability is one among these manifestations of genomic instability (Chaudhuri and Chaudhuri, 1994).
It corresponds to an alteration in size of simple repeat sequences, such as di- or trinucleotide repeats. These sequences are common near to gene or within gene loci. A finding of SSR-instability implies the presence of changes in at least one gene involved in inducing metabolic diseases (Thibodeau, et al. 1993; Markowitz et al. 1995).
However, the whole procedure from the construction of a genomic library to the synthesis of specific primers from the flanking sequences is time-consuming and expensive. Then, a variant of the SSRs technique has been described where microsatellite oligonucleotides amplify genomic segments different from the repeat region itself.
This approach, named Inter-SSR (ISSR), employs olignucleotides based on a simple sequence repeat anchored at their 5′- or 3′- end by two to four arbitrarily chosen nucleotides. This triggers site-specific annealing and initiates PCR amplification of genomic segments, which are flanked by inversely oriented and closely spaced repeat sequences.
During the mid 1990s, DNA chips with microarrays of DNA samples became available to achieve very fast speeds in generating information about DNA sequences (Gupta et al. 1999). This considered a major technological breakthrough in the field of DNA monitoring protocols and that can be compared with the role of semiconductors in the field of electronics.
The microarrays are prepared on solid glass (silica) plates where high-density single stranded 20 mers DNA probes (often cDNAs) are layered. These microarrays are hybridised with an unknown labelled DNA sample, and the hybridization patterns are analyzed with computer device.
With this protocol, a single experiment can reveal several thousand DNA-DNA hybridisation reactions. Popular amongst them are— RFLP, RAPD, SSR, AFLP, and SNP (single nucleotide polymorphism). The major drawback of early PCR techniques is their sensitivity to DNA quality or reaction conditions. However, SNP and AFLP could resolve these difficulties to a large extent.
Use of Molecular (DNA) Markers to Detect Plant Diversity:
The methods for molecular biology provide a high-resolution view of phylogenetic differences among individual plants at both intra-and the interspecific levels. These high resolution data can be used to address a number of questions that arise in plant diversity.
Some of the Important Questions that are Currently being Addressed are:
(1) what rates of nucleotide substation are characteristic of plant genes?
(2) What can be deduced about plant relationships from DNA sequence data?
(3) From DNA data what can be inferred about gene diversity in plants?
(4) What can be inferred about the functional constraints on molecules from the pattern of DNA sequence divergence ?
Since mid-1980s, large numbers of data are available about the divergence of chloroplast encoded genes (Rubisco L), chloroplast ribosomal proteins, and chloroplast ATPase genes (Zurawski and Clegg, 1987). Being prokaryotic genome in nature, unlike plant nuclear genome, chloroplast DNA has minimum amount of repetitive DNA elements. This has made the study less problematic.
Later, sequence divergence studies in maize transposable elements (Ac and Ds) showed that Ds 1 family underwent an expansion in numbers during a restricted period of time in the genome of the maize progenitor and Tripsacum. Since then molecular data had emerged from plant-pathogen interaction. Specificity between host and pathogen is determined by a clear gene-for-gene interaction.
For example, polymorphic combinations of probe and endonucleases could pin point pathogen loci in genetic map of lettuce and the linkage map of lettuce mildew could be established (Michlemore et al. 1989). These findings were based on RFLP analyses. Later, PCR based technology was successfully applied for genetic analysis of several plant species.
The RAPD technology has been introduced to genetic mapping of woody plants, gymnosperms and angiosperms (Carlson et al. 1991; Bucci and Menozzi, 1993; Koller et al. 1993; Dunemann. 1994; Weeden et al. 1994) where it could be inferred that level of heterozygosity is generally high in douglas fir and spurce. Reiter et al. (1992) applied RAPD technology, to generate linkage in Arabidopsis.
Later, many other authors applied this PCR technique in many angiosperm species and crop plants (Mulcahy et al. 1992; Tinker et al .993. Yu and Pauls, 1993a, 1993b). Bandopadhyay (2003) and Roy (2003) have reviewed the subject.
Use of SSR markers for plant genomes were made for the first time in 1991. However, Akkaya et a; 1992 was the first to use its successful application in soybean genomes. Later, many paint molecular biologists delete reported diversity in different plant species using PCR technology. Microsatellite loci, however differ from RFLPs since homeoloci are not always available (Schlotterer, 1998; Vendramin et al. 1999). Ma and Lapitan (1998), and many other workers could successfully use AFLP markers in plant genetics, e.g. in rice, barley, wheat etc.
Today, there is no single classification for the Iris genus. Using the RAPD-method, the same authors typified the DNA of representatives of the Russian Far Eastern irises. Each of the iris species has its specific spectrum of RAPD-products characterized by a specific set of fragments.
F. E. Branch, Russian Academy of Sciences, uses RAPD-analysis to conduct population genetic studies of rare plant species that need protection, as well as widespread species involving complex taxonomic issues, whose resolution would help clarify the evolutional history of species. Russia’s Maritime Province is the only place on earth where real ginseng (Panax ginseng C. A. Meyer) is preserved in natural habitats.
An examination of the extent of genetic variability of natural populations should be the theoretical foundation for developing a scientifically grounded program for preserving ginseng. The molecular markers could identify species, natural populations and cultivated forms of real ginseng, and also continental and insular populations of irises and Larchwood populations.
Using several arbitrary primers, the DNA of representatives from three natural ginseng populations originating in several Russian Districts were studied. The populations showed variations in genetic make up, which often differed not only in the presence of polymorphous loci in the DNA of several plants, but in varied intensity of homologous fragments in DNA amplification profiles.
The institute’s workers also investigated ginseng forms cultivated in nature and in vitro. They obtained results which showed that cultivation in vitro leads to substantial changes in the DNA. Similar works were reported with many other Indian or non-Indian plant species. While working on castor plant (Ricinus communis L.) genotypes, it was found that the established breeding lines differ significantly in PCR amplification and a few markers can successfully fingerprint them.