In this article we will discuss about the structure and components of bacterial cell.
Structure of Bacterial Cell:
Bacterial cells (prokaryotic cells) are structurally much simpler than eukaryotic cells and the two cell types are compared in Table 3.2. They consists of various cell surface structures, cell wall, plasma membrane, many cytoplasmic inclusions, and the bacterial chromosome (nucleoid). Except some, all structures do not occur in every genus.
Furthermore, gram-negative and gram-positive bacteria differ, particularly, with respect to their cell walls. Despite these variations, however, the bacterial cells are consistent in their fundamental structure and most important constituents.
Bacterial cells almost always are surrounded by a chemically complex cell wall. Plasma membrane lies inside the cell wall and is separated from the latter by a periplasmic space. This membrane can be invaginated to form simple internal membranous structures called mesosomes.
Since the bacterial cells lack internal membrane-bound organelles, they appear morphologically simple internally. The genetic material is localized in a discrete region called bacterial chromosome or nucleoid, and in the form of extra-chromosomal genetic material referred to as plasmids. The latter is not separated from the surrounding cytoplasm by nuclear membrane or any other membrane.
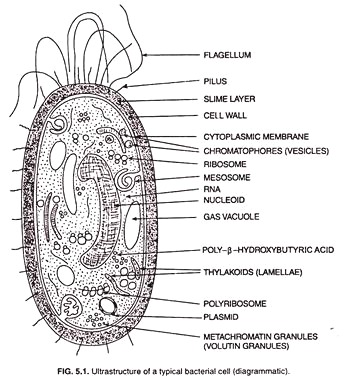
Ribosomes and distinct masses called cytoplasmic inclusions are scattered in the cytoplasmic matrix. Both gram-positive and gram-negative bacteria cells possess flagella for locomotion. In addition, certain type of bacterial cells possess small hair-like structures and the glycocalyx (slime layer, capsule) external to their cell wall. However, the subcellular organization of a typical bacterial cell in shown in Fig. 5.1.
The ultra-structural details and the functions of the subcellular structures of a bacterial cell are taken at length.
Components External to Cell Wall of Bacterial Cell:
Bacterial cells possess various structures external to the cell wall that basically contribute in protection attachment to objects, and cell movement.
The Glycocalyx: Slime Layer and Capsule:
The glycocalyx represents the gelatinous covering around many bacterial cells secreted at the time of their active growth. This covering consists of polysaccharides of high molecular weight (slime layers and capsules) and, in few cases, of proteins (S-Iayers). When the glycocalyx does not form a persistent layer, but is present more diffusedly forming a loose mass around the bacterial cell, it is called slime layer.
Slime layer is more easily deformed, is more difficult to see, and can be very easily removed by washing the bacterial cells. Gliding bacteria often produce slime, which presumably aids in their motility. The slime probably attaches them to the substratum and lubricates the surface for more efficient movement. When the gelatinous covering forms a well- defined persistent layer, it is called capsule.
Capsules are composed generally of polysaccharides (e.g., Streptococcus mutans, S. salivarious, Xanthomonas, Corynebacterium) that contain, apart from glucose, aminosugars, rhamnose, uronic acids of various sugars, 2-keto-3- deoxygalactonic acid, and organic acids such as pyruvic acid and acetic acid.
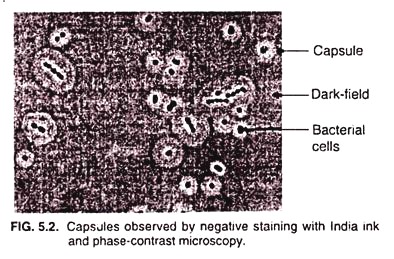
However, the capsules of some bacilli bacteria (e.g., Bucillus subtilis, B. anthracis) consist of polypeptides, mainly poly-D-glutamic acid. Capsules can be seen by negative staining with dyes that do not penetrate the capsular material, such as India ink, Chinese ink, Nigrosin or Congo red. In negative staining, the capsule appears light against a dark background (Fig. 5.2).
Although capsules are not required for growth and reproduction of bacterial cells in vitro, they do confer several advantages when bacteria grow in their natural habitats.
Capsules however perform several functions the important ones of which are the following:
(i) They help attaching bacterial cells on diverse surfaces (e.g., plant root surface, human teeth and tissue rocks in fast flowing streams) because of its sticky nature. Pathogenic bacteria that enter the human body by specific routes usually do so by first binding specifically to surface components of host tissues.
(ii) Capsules protect bacterial cells against phagocytosis by various protozoans and WBCs and, therefore, capsulated bacteria pathogens are more virulent. For example, capsulated Streptococcus pneumonia strains cause pneumonia, while non-capsulated ones are phagocytized.
(iii) Capsules protect bacterial cells from desiccation. Since the outer polysaccharide layers of capsule are hygroscopic and bind a significant account of water, it is considered that these layers contribute some role in resistance to desiccation.
(iv) Some bacteria (e.g., Streptococcus mutans) use its capsule as source of energy by breaking down the sugars of capsule when stored energy content is considerably reduced during adverse conditions.
(v) Capsules help inhibit the movements of nutrients from the bacterial cell.
(vi) Capsules prevent bacterial viruses (bacteriophages) and most hydrophobic toxic materials (e.g., detergents) from attaching on to bacterial surfaces.
Paracrystalline Surface Layers (S-Layers):
Many gram-positive and gram-negative bacteria possess a regularly structured surface layers called S-layers, on their surface. S-layers are widespread among archaebacteria where they may be the only wall structure outside the plasma membrane.
S-layers are composed of a two-dimensional array of protein or glycoprotein and have a crystalline appearance accompanied with various symmetries such as tetragonal, hexagonal, or trimeric depending upon the number and structure of the protein or glycoprotein subunits (Fig. 5.3).
In gram- native bacteria the S-layers adhere directly to the outer membrane, whereas they are associated with the peptidoglycan surface in gram-positive bacteria.
Although the major function of S-layers is not known, they are considered to perform the following functions:
(i) Being interface between the bacterial cell and its environment, the S-layers are thought to act as a selective seive which allows the passage of low molecular weight substances and excludes large molecules and structures (e.g., viruses). By this act, they help retain proteins near the bacterial cell much like the outer membrane does in gram-negative bacteria.
(ii) S-layer may protect bacterial cell against ion and pH fluctuations, osmotic stress, enzymes, or the predaceous bacterium Bdellovibrio.
(iii) S-layers help maintaining the shape and envelop rigidity of bacterial cells that possess it, and can promote cell adhesion on to various surfaces.
(iv) Evidences suggest that the S-layers of pathogenic bacteria that contain them are protected against certain host defence mechanisms such as complement attack and phagocytosis.
Fimbriae, Pili and Spinae:
Bacterial cells possess short, fine, hair like, protein-contributed appendages that extend from the cell surface. These appendages are called fimbriae (ring. fimbria) and pili (sing, pilus), are thinner than flagella, and are not involved in locomotory activities. The term ‘pili’ was introduced by Brinton (1950) and ‘fimbriae’ by Duguid et al. (1955).
Although many people use the term ‘fimbriae’ and ‘pili’ interchangeably, recent evidences suggest that these two should be considered separate at least at the level of sex and be recognized as fimbriae and sex pili.
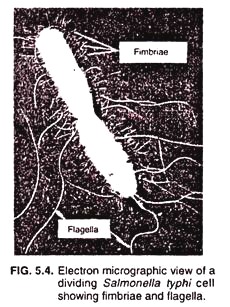
Fimbriae (Fig. 5.4) are considerably shorter than sex-pili and are more numerous; a single cell may be covered with upto 1,000 fimbriae. The latter can only be seen in an electron microscope due to their small size. Each fimbria is about several micro-meters long and about 3 to 10 nm in diameter, and is composed of helically arranged protein subunits.
The protein that constitutes the fimbriae is called pilin, the molecular weight of which being about 17,000 Da. Many classes of fimbriae are recognized on the basis of their structure and function.
The fimbriae belonging to class IV, called type IV fimbri/pili, is involved in an unusual form of motility in certain bacteria. This motility is called twitching motility, which is a type of movement on solid surfaces where rapid and reversible extension and retraction of the fimbriae allow the bacterial cell to crawl along the surface. Unlike fimbriae of other classes, type IV fimbriae occur only at the poles of bacterial cells.
Important functions performed by fimbriae are the following:
(i) Certain types of fimbriae attach bacteria to solid surface such as rocks in streams or host tissues,
(ii) In case of certain pathogenic bacteria including Salmonella typhimurium (Salmonellosis), Bordetella pertussis (whooping cough), and Neisseria gonorrhoeae (gonorrhea), the fimbriae enable bacterial cells to form pellicles or biofilms on tissue surfaces.
(iii) As stated above, type IV fimbriae are required for the twitching mobility that occurs in bacteria such as Pseudomonas aeruginosa. N. gonorrhoeae, and certain strains of E. coli. These fimbriae also appear to be involved in some sorts of gliding mobility by myxobacteria.
(iv) Recent evidences suggest that the type IV fimbriae mediate the transfer of genetic material from one bacterial cell to another in a wide variety of bacteria by the process of transformation.
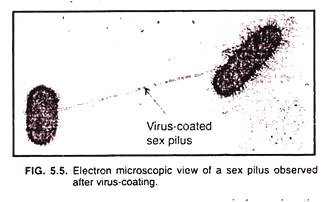
Sex pili (sing, sex pilus) are structurally similar to fimbriae but arc longer than them, and are 9 to 10 nm in diameter. Only one to a few sex pili are reported on bacterial cell surface. Since the sex pili serve as receptors for certain types of viruses, they can be observed under the electron microscope when they become coated with virus particles. (Fig. 5.5).
Sex pili are genetically determined by sex factors or conjugative plasmids and are clearly involved in the process of conjugation, a process of transfer of genetic material from one bacterial cell to another. If sex pili are absent or if the bridge established by sex pili between two bacterial cells is interrupted, the conjugation process is not completed.
Spinae have been reported in some gram-positive bacteria. These are tabular, pericellular, and non- prosthecate rigid hairy appendages said to help adjust bacterial cells to some environmental conditions such as pH, salinity, temperature, etc. Spinae are made of a single protein, namely, spinin.
Flagella (sing. Flagellum):
Bacterial flagella are long (upto 15-20 µm) and thin (about 20 nm) appendages free at one end and attached to the cell at other end. They are the organs of locomotion and are so thin that a single flagellum can only be observed with a light microscope after staining with special flagella-stains that increase their diameter.
Flagella can easily be seen with the electron microscope. Bacterial flagella arise from the plasma membrane and pass out through the cell wall.
Structure:
A bacterial flagellum is not straight but helical and consists of three distinct parts—the filament, the hook and the basal body (Fig. 5.6). The filament is the thinner, elongated, terminal part attached to the hook; the hook represents a somewhat broader and thicker basal region that passes out through the cell wall; and the basal body constitutes the extreme basal part attached to the plasma membrane.
1. Filament:
The filament is a fine, cylindrical, helical hollow structure, about 120-200 Å in diameter. It is composed of a fibrin protein called ‘flagellin’ structurally similar to keratin and myosin proteins and ranging in mol. Wt. from 30,000 to 60,000 dalton. A cross section of the filament reveals that there are eight flagellin molecules surrounding a central hollow cylinder.
Actually, the 8 flagellin molecules seen in the cross section arc the parts of flagellin molecule-chain eight in number and running longitudinally around the central hollow cylinder.
Each chain contains approximately 1,000 spherical, smaller flagellin molecules each of 40 Å diameter. In this way, the bacterial flagellum fundamentally differs from the flagellum of an eukaryotic cell, which has 9 + 2 type of arrangement in its filament.
2. Hook:
As said earlier, hook is a somewhat broader and thicker basal region of a flagellum and passes out through the cell wall. It is made up of a single type of protein and functions to connect the filament to the basal body of the flagellum.
3. Basal Body (The Motor Portion of the Flagellum):
Basal body, the motor portion of the flagellum is the most complex part of a flagellum (Fig. 5.7). In gram-negative bacteria, the basal body has five rings (L, P, S, M, and C) connected to a central rod.
The outer L (lipopolysaccharide) and P (peptidoglycan) rings remain embedded in the lipopolysaccharide and the peptidoglycan layers, respectively; the inner S (supermembrane) and M (membrane) rings are located within the plasma membrane, and the innermost C (cytoplasmic) ring is located in the cytoplasm.
In gram-positive bacteria, which lack the outer lipopolysaccharide layer, only S, M and C rings are present. There are a pair of proteins called Mot A and Mot B that surround the inner ring and are associated to the plasma membrane. In addition to Mot proteins, there is a final set of another proteins called Fli proteins (Fli G, Fli M, and Fli N) associated with the C ring located in the cytoplasm.
Actually the portion of the basal body that rotates (the motor) is consisted of the rod, the M-ring, and the Mot and Fli proteins. The Mot proteins drive the motor causing a torque that rotates the filament, whereas the Fli proteins function as the ‘motor switch’ reversing rotation of the flagella in response to signals send by the bacterial cell.
Flagellar Movement:
Bacterial (prokaryotic) flagella operate in different manner when compared to eukaryotic flagella. Each individual flagellum is a semi-rigid helix and moves by rotation like propellers on a boat (Fig. 5.8). Monotrichous and lophotrichous polar flagella rotate counter-clockwise and make the bacterial cell spring around and run forward from place to place with the flagellum trailing behind.
These bacteria stop and tumble randomly by reversing the direction of flagellar rotation. The flagella of peritrichous bacteria rotate counter clockwise, like montrichous and lophotrichous ones, to move forward. The flagella bend at their hooks to form a rotating bundle that propels them forward. Clockwise rotation of the flagella disrupts the bundle and the cell tumbles.
Flagellar Movement Mechanism:
The rotatory motion of the flagellum is imparted from the basal body, the motor attached at the base. A rod or shaft extends from the hook and ends in the M-ring which can rotate freely in the plasma membrane.
Though the exact mechanism of flagellar rotation still is not clear, it is believed that the S-ring is attached to the cell wall in gram-positive bacteria cell and does not rotate. The P and L-rings of gram-negative bacteria act as bearings for the rotating rod.
The energy required for the rotation of the flagellum comes from the proton motive force (PMF), not directly by ATP as is the case with eukaryotic flagella. The plasma membrane becomes energetically charged and this energized state of plasma membrane is called proton motive force (PMF).
During this energized state of plasma membrane the protons (H+) are separated from hydroxyl ions (OH–) and are translocated across the plasma membrane through Mot proteins. Proton (H+) translocation across the plasma membrane through Mot proteins exerts electrostatic forces on charges present on the C, N, and S rings resulting in attractions between positive and negative charges.
This attraction between positive and negative charges causes the basal body to rotate. Calculations have shown that about 1000 protons must be translocated per single rotation of the flagellum.
Flagella do not rotate at a constant speed but instead can increase or decrease their rotational speed in relation to the strength of the proton motive force. Flagellar rotation can move bacteria through liquid media at speeds of up to 60 cell lengths/sec.
Although this is only about 0.00017 km/h, when comparing this speed with that of higher organisms in terms of number of lengths moved per second, it is extremely fast. By comparison, the cheetah, the fastest land animal, can sprint at 110 km/h. which is approximately 25 body length/sec.