The following points highlight the top three types of non-Genetic ribonucleic acid (RNA). The types are: 1. Messenger RNA (MRNA) 2. Ribosomal RNA (rRNA) 3. Transfer RNA (tRNA).
Type # 1. Messenger RNA (MRNA):
Monod and Jacob (1961) coined the term ‘messenger RNA’ to describe the template RNA that carries genetic information for protein synthesis from nuclear DNA to ribosomes in the cytoplasm. Messenger RNA (mRNA) makes up less than 5% of the total cellular RNA.
The molecular weight of an average sized mRNA molecule is about 500000 and its sedimentation coefficient is 8S; mRNA varies greatly in length and molecular weight.
Structure:
mRNA is always single-stranded. It contains mostly the bases adenine, guanine, cytosine and uracil. There are a few unusual substitute bases also. Although there is a certain amount of random coiling in extracted mRNA, but there is no base-pairing. Usually each gene transcribes its own mRNA. Therefore, there are approximately as many types of mRNA molecules as there are genes.
The eukaryotic mRNA molecule has the following structural features (Fig. 4.30):
(i) Cap:
At the 5′ end of the mRNA molecule, in most eukaryote cells and animal virus is found a ‘cap’. This is blocked methylated structure, m7Gpp Nmp Np or mZGpp Nmp Nmp Np, where N = any of the four nucleotides and Nmp = 2’0 methyl ribose. The rate of protein synthesis depends upon the presence of cap. Without the cap mRNA molecules bind very poorly to the ribosomes.
(ii) Non Coding Region 1 (NC 1):
The cap is followed by a region of 10 to 100 nucleotides. This region is rich in A and U residues, and does not translate protein.
(iii) Initiation Codon is AUG in both prokaryotes and eukaryotes.
(iv) The coding region consists of about 1500 nucleotides on the average and translates protein.
(v) Termination codon:
Termination of translation on mRNA is brought about by a termination codon. In eukaryotes the termination codons are UAA, UAG or UGA.
(vi) Non-coding region 2 (NC 2):
Consists of 50-150 nucleotides and does not translate protein. This region contains an AAUAAA sequence in all cases.
(vii) Poly (A) sequence:
At the 3′ end is a polyadenylate or poly (A) sequence, which initially consists of 200-250 nucleotides, but becomes shorter with age. This poly (A) sequence is added in the messenger as a tail before the mRNA reaches the cytoplasm. Messenger RNA combines with ribosomes to form polyribosomes or polysomes which serve as work-benches for protein synthesis. Each polysome contains several ribosomes.
Transcription and Processing:
The mRNA is initially transcribed as a precursor called pre-mRNA otherwise termed as primary transcript subsequently shortened by splicing. The 5′ end of the mRNA is modified by capping and the 3′ end-is modified by polyadenylation. RNA transcribed by RNA polymerase II exists in the nucleus as a population of molecules known as heterogeneous nuclear RNA (hn RNA).
Splicing:
This process involves the removal of non-essential sequences from the primary transcript of each gene. Each gene is made up of essential and non-essential sequences, i.e., exons and introns, alienating with each other. The non-essential sequences of primary transcript corresponding to introns are scissored off by specific enzymes and the essential sequences are joined end to end. This entire process is termed splicing.
Capping:
Eukaryotic mRNAs are modified at the 5′ end by addition of the modified nucleotide, 7-methylguanosine, in an unusual
5′ →5′ triphosphate linkage to the first nucleotide of the mRNA.
Polyadenylation:
Most eukaryotic mRNAs are modified at their 3′ ends by the addition of a poly A tail.
Function:
mRNA acts as a template for protein synthesis. It is produced in the nucleus by transcription of protein coding genes with the help of RNA polymerase.
Type # 2. Ribosomal RNA (rRNA):
Ribosomal RNA (rRNA) or insoluble RNA constitutes the large part (up to 80%) of the total cellular RNA. It contains the four major RNA bases with a slight degree of methylation and shows differences in the relative properties of the bases between species. The molecules appear to be single polynucleotide strands which are branched and flexible.
At low ionic strength rRNA behaves as a random coil, but with increasing ionic strength the molecule shows helical regions, produced by base-pairing between adenine and uracil; guanine and cytosine.
Types:
The eukaryotic cell have four kinds of rRNA molecules (Fig. 4.31):
(i) 28S rRNA,
(ii) 18S rRNA,
(iii) 5S rRNA,
(iv) 5.8S rRNA.
The 28S rRNA, 5.8S rRNA and 5S rRNA occur in 60S ribosomal subunit while 18S rRNA occurs in 40S ribosomal subunit of SOS ribosomes of eukaryotes.
The prokaryotic cells contain three kinds of rRNA molecules (Fig. 4.31):
(i) 23S rRNA (occur in 50S ribosomal subunit),
(ii) 16S rRNA (occur in 30S ribosomal subunit),
(iii) 5S rRNA (occur in 50S ribosomal subunit)
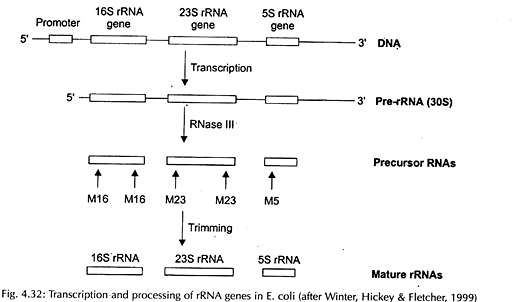
Transcription and Processing:
Transcription and processing of rRNA in prokaryote and eukaryote have been represented in Figs. 4.32 and 4.33 respectively.
Function:
The rRNA performs several functions:
(i) Gives structural integrity to ribosome;
(ii) Serves as the site for mRNA;
(iii) Has a role in protein synthesis.
Type # 3. Transfer RNA (tRNA):
The RNA which possesses the capacity to combine specially with only one amino acid in a reaction mediated by a set of amino acid specific enzymes called aminoacyl-tRNA synthetase, transfers that amino acid from the ‘amino-acid pool’ to the site of protein synthesis and recognizes the codons of the mRNA, is known as the transfer RNA (tRNA) or soluble RNA (sRNA).
Structure:
These are the smallest molecules of RNA. Each tRNA has an anticodon of three nucleotides. Each tRNA has a molecular weight of about 30000 and the sedimentation coefficient of mature eukaryote tRNA is 3.8S. It is made up of 73-93 nucleotides. The tRNA molecule consists of a single strand looped about itself.
The nucleotide sequence (primary structure) of tRNA was first worked out by Holley et al. (1965) for yeast alanine tRNA. Since then the sequence of different tRNAs, ranging from bacteria to mammals, has been established.
Some modified bases in tRNA as nucleosides are inosine (hypoxanthine), pseudouridine, dihydrouridine, 7-methylguanosine, etc. R. Holley proposed a secondary structure of tRNA “the clover- leaf model” which is the most widely accepted structure. The structural features of this two- dimensional tRNA structures are as follows (Fig. 4.34).
The two most essential components responsible for recognizing amino acid on the one hand and the mRNA on the other are:
(i) Acceptor end and
(ii) Anticodon loop.
(i) The Acceptor End:
All tRNA molecules contain the same terminal sequence of 5′ CCA 3′ bases at 3′ end of the polynucleotide chain. The last residue, adenylic acid (A) is the amino acid attachment site. In addition, it contains other accessory loops or arms.
(ii) The Anticodon Loop:
The anticodon loop is directly opposite to the acceptor end having three bases to recognise and form hydrogen bonds with the mRNA, thus reading the genetic message. It is complementary to the corresponding triplet codon of mRNA.
(iii) The D-Loop (Di-hydro-uridine loop) or DHU arm:
It is made up for the recognition by the specific aminoacyl tRNA synthetase and is made up of 8-12 unpaired bases.
(iv) The t-loop (TΨC arm) or ribosomal binding loop: it is thought to interact with a complementary region of 5S ribosomal RNA during protein synthesis. It is involved in the binding of tRNA molecules to the ribosomes. It is made up of seven unpaired bases including pseudouridine (Ψ) so it is known as TΨC arm.
(v) The extra, optional or variable arm:
It occurs only in some tRNAs. It may be small containing only 2-3 nucleotides (class I tRNAs) or larger containing 13-21 nucleotides with up to five base pairs in a stem (class II tRNAs).
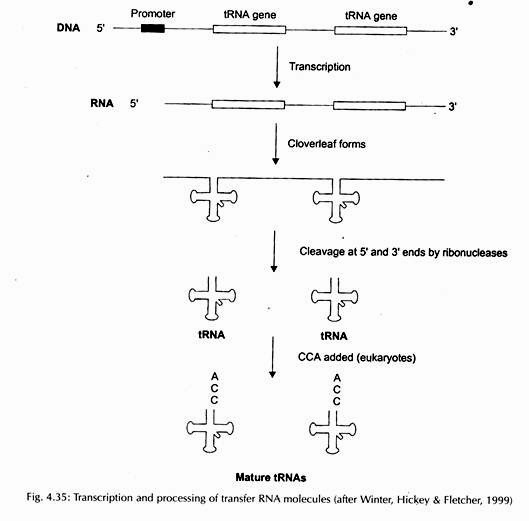
Transcription and Processing:
Transcription and processing of tRNA have been presented in Fig. 4.35.
Function:
It plays a key role in protein synthesis, carrying amino acids to the site of protein synthesis and attaches itself to ribosome in accordance with base sequence specified in mRNA (codon) by their anticodon. The crucial factor in enzyme-protein synthesis from hereditary messenger carried by the mRNA is the codon-anticodon relationship in translation.
The anticodon located in tRNA in the form of 3 bases, must be complementary with the triplet codon in the messenger RNA. As such this sequence complementarity between the two is the most important event of enzyme-protein synthesis.