This article throws light upon the three innovative methods used in molecular biology.
The three innovative methods used in molecular biology are: (1) Cell Fractionation (2) Nucleic Acid Purification, Yield Analysis of Purified DNA and (3) DNA Sequencing.
Contents
Method # 1. Cell Fractionation:
Organelles are membrane-enclosed vesicles inside all eukaryotic cells that function in a variety of important cellular processes. A method is described here for isolation of various sub-cellular fractions from rat liver by a technique termed differential centrifugation. These fractions can be used for assay for the presence of specific organelles using enzyme assays.
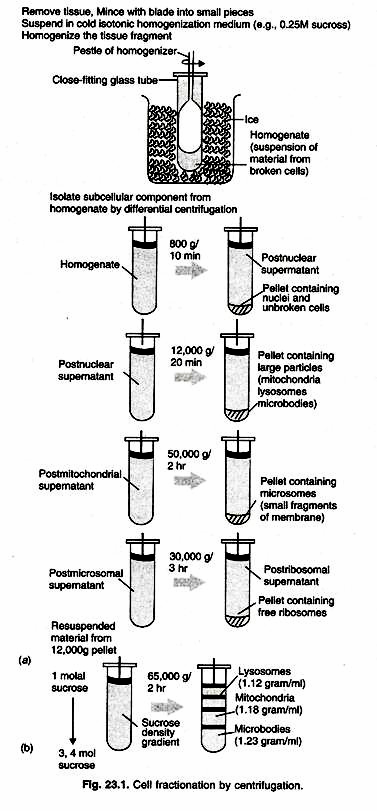
The arrangement of macromolecules within a cell is as important to cellular function as their catalytic activities. Cellular compartmentalization provides efficiency by bringing together related compounds that can interfere with each other (i.e., lysosomal hydrolytic enzymes). Cellular compartmentalization is accomplished in part by various sub-cellular organelles. The method utilizes differential centrifugation to isolate components of different densities. With this technique, the heaviest or most dense organelles, nuclei pellet in less time and with less force than is required to pellet lighter organelles such as mitochondria. First, a cell homogenate is made by rupturing the cell membranes in the tissue.
The homogenate is then centrifuged for a short period of time to remove cell debris and nuclei. The supernatant is then transferred to another tube and centrifuged longer to pellet the lighter mitochondria. The method of fractionation depends upon tissue type and organelle to be isolated. Homogeneous cell populations from cell culture are well suited for cell fractionation. Some tissues, like those in the liver also have one cell type that predominates, so are also well suited. Most chlorophyll-free plant tissues are acceptable for preparing mitochondria, but recently-harvested plant tissues are usually required.
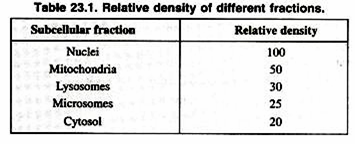
Once a cell type is chosen, it is important to obtain the organelles in a biochemically active, morphologically whole state (Table 23.1). Homogenizers are used to break open the cells without damaging the organelles. Homogenizers have a precise clearance between the glass tube and pestle, which breaks the cell membrane leaving the smaller organelle membranes intact. The homogenization buffer is a solution which often includes sucrose to partially dehydrate the organelles, keeping them intact.
No technique used to isolate organelles is perfect. It is very difficult to get pure unbroken preparations of any organelle. Techniques providing optimal isolation of one organelle may completely rupture another organelle. Thus methods are often used to measure the contamination of one organelle fraction by another. This can be done by analyzing each organelle fraction for organelle-specific marker enzymes.
Method of Tissue Homogenization:
1. Transfer about one-half (~2 gm) of the tissue to a glass homogenizer on ice. Do not use a tight-fitting ground-glass homogenizer in this step since the organelle membranes may disrupt partially.
2. Add 18 ml of ice-cold homogenization buffer to the homogenizer. Note: the Calcium in the buffer is used here to stabilize the organelle membranes.
3. Homogenize 3-5 strokes until a homogeneous homogenate is made. Keep the homogenizer on ice. Caution: Excessive grinding or heating can damage and inactivate sub-cellular fractions.
4. Transfer the homogenate into a plastic 50 ml tube on ice.
5. Repeat steps 6-9 (combining all homogenates) until all the minced liver tissue has been homogenized (Fig. 23.1).
Isolation of Nuclei:
6. Centrifuge the tube at 700 x g for 10 min at 0°C. The pellet contains the cell debris and nuclei. The supernatant contains all the lighter cellular fractions.
7. Without disturbing the crude nuclear pellet, carefully decant the supernatant into another tube on ice. Since the mitochondria are present in supernatant, this is used for the Isolation of the Mitochondria, while pellet can be used for the isolation of the nuclei.
8. To continue with the nuclear isolation: Nuclear Wash:
a. Add 10 ml of ice-cold homogenization buffer to your crude nuclear pellet.
b. Use a teflon homogenizer to re-suspend the pellet in the buffer.
c. Place two layers of cheesecloth in a funnel over a tube. Filter the 10 ml suspension through the cheesecloth.
d. Rinse the debris in the filter with another 10 ml of homogenization buffer collecting the filtrate in the tube. Discard the filter with its cellular debris.
e. Centrifuge the combined filtrates (20 ml) at 1500 x g for 10 min at 0°C. The pellet contains the washed nuclear fraction. Without disturbing the nuclear pellet, discard the supernatant.
9. Add 5.0 ml of ice-cold homogenization buffer to the nuclear pellet.
10. Use a teflon homogenizer to re-suspend the pellet in the buffer.
11. Using a pipette, aliquot the nuclear suspension into two glass test tubes.
12. Label the two glass tubes “Nuclei”, and store them at -20 °C. Caution: Glass tubes should not be greater than half full, or they may crack upon freezing.
Isolation of Mitochondria:
13. Obtain the supernatant from step 12 in a tube on ice.
14. Centrifuge at 5,000 x g for 15 min at 0°C. The pellet contains the crude mitochondrial fraction. The supernatant contains microsomes, membrane fragments, ribosomes, cytoplasmic enzymes, etc.
15. Without disturbing the mitochondrial pellet, carefully decant the post-mitochondrial supernatant (and the pink partially sedimenting layer) into a graduated cylinder.
16. Pour the supernatant into a clean tube on ice.
17. The supernatant can be used for the “Isolation of the Microsomes”.
18. To continue with mitochondrial isolation: Mitochondrial Wash;
a. Add 20 ml of ice-cold homogenization buffer to the crude mitochondrial pellet.
b. Using a teflon homogenizer, resuspend the pellet in the buffer.
c. Pour the suspension back into the tube, add another 20 ml of buffer, mix, and centrifuge at 234,000 x g for 10 min at 0°C. The pellet contains the washed mitochondria.
d. Without disturbing the mitochondrial pellet, decant and discard the supernatant.
19. Add 5.0 ml of ice-cold homogenization buffer to the mitochondrial pellet.
20. Using a teflon homogenizer, re-suspend the pellet in the buffer. This is how using differential centrifugation, various organelles can be isolated and studied.
Method # 2. Nucleic Acid Purification, Yield Analysis of Purified DNA:
Purification Genomic and Plasmid DNA:
Genomic and plasmid DNA can be isolated from cells by disrupting cells and then exploiting the differences in properties between DNA, protein and other constituents. There is no universal method for purification of genomic DNA. The method for purification of DNA depends on the nature of the source material. DNA from plants is more difficult to obtain in clonable form than from animals due to presence of secondary metabolites and polysaccharides and the extent of interfering substances present in that source.
The first reported purification of chromosomal DNA was in 1944 by Avery and co-workers from the “S” cells of Diplococcus pneumoniae Type II. These cells contained a very large amount of capsular polysaccharide and the method was developed to purify the DNA and remove the contaminant.
From about fifteen years Avery’s method with some modification continued to be used for DNA solution. It was gradually realized that there was need to improve upon the yield and also the size of DNA. The deoxynucleases that were released on cell lysis degraded a large part of DNA; Marmur introduced the use of alkaline buffer (0.1 M Tris, pH 9.0) and sodium dodecyl sulphate. Both the high pH and sodium dodecyl sulphate were found to inactivate the nucleases.
In addition, lysis operation to be performed at low temperature (0-2 °C) to slow down the enzymes and the subsequent extraction with chilled phenol saturated with the pH 9.0 buffer, which denatured the proteins bound to DNA and consequently increased the yield of DNA.
The DNA was precipitated from the aqueous layer with ethanol as was done by Avery. No protein could be detected by the Lowry test in the DNA sample, but some RNA was present. All methods subsequently developed for isolation of chromosomal DNA from bacteria, viruses and animal cells in culture have been largely derived from the method of Marmur. EDTA has been introduced in the lysis medium to inactivate nucleases which are known to be dependent on Mg++. In some methods phenol has been replaced by a mixture of equal volumes of phenol and chloroform.
Since phenol treatment is thought to shear large DNA molecules, methods have been developed avoiding phenol and such methods probably yield better quality DNA at the expense of quantity. When DNA is desired to be isolated from animal or plant tissue, the above mentioned steps have to be preceded by procedure to obtain cell powder from the ground tissue. The tissue is minced finely and then disintegrated thoroughly in a Warring blender with liquid nitrogen. This step is sometimes performed in a mortar and pestle. The powder left behind after evaporation of the liquid nitrogen is extracted with the alkaline buffer (pH 8.0 to 9.0) containing EDTA and the detergent.
Subsequently the mixture is treated with protease and ribonuclease followed by phenolisation. A problem that is quite often encountered with plant tissue is the presence of a large amount of polysaccharides. Plant DNA free from polysaccharides is obtained by cesium chloride ethidium bromide equilibrium centrifugation as described under purification of plasmid DNA.
Different Types of Isolation Procedures:
1. SDS-phenol Extraction:
This is the simplest and fastest method for isolation of DNA and is widely used with filamentous fungi and plant material. The technique uses SDS with phenol to denature and dissolve macerated hyphae leaving the DNA intact. DNA is then precipitated from solution with a precise volume of isopropanol.
2. SDS-proteinase K Treatment:
This technique is applicable to most materials and relies on proteinase K and SDS to dissolve the sample and digest the protein component without affecting the DNA. The sample is then extracted with protein denaturants (phenol, phenol-chloroform, chloroform) and the aqueous liquid containing the DNA is dialyzed and precipitated with ethanol or isopropanol in sodium or ammonium acetate solution. RNAse may or may not be used depending on different protocols used.
3. CTAB Treatment:
Isolation of DNA from plants is preferably achieved by treatment with CTAB. This detergent carries a positive charge which interacts with the negative charge of DNA in high salt concentration and forms a soluble complex. In subsequent steps a decrease in the salt concentration causes precipitation of DNA, leaving other compounds, especially polysaccharides, in solution. The preparation of DNA from animal tissue which is in mucopolysaccharides could also be done by this technique which yields good quality material.
Plasmid DNA Isolation:
Plasmids are extra chromosomal double stranded circular DNA molecules present in bacteria and lower eukaryotes which exist in super helical state in vivo, the size of plasmid being in the range between 1 kb and more than 300 kb. Chromosomal DNA is much larger and though could also be circular is made linear by shear pressure during cell lysis and the DNA isolation procedure. These long linear molecules invariably get entangled in the debris of lysed cells.
The early procedures of plasmid isolation were developed in the late 1960s in the laboratory of Donald R Heiliski in the University of California at San Diego. Cell lysis was achieved by using EDTA, lysozyme and mild non ionic detergent like Triton X-100 at pH 8.0 and avoiding vigorous shaking. A centrifugal step was then incorporated to preferentially sediment the chromosomal DNA molecules entangled in the cellular debris.
The supernatant solution contained the plasmids, which was treated successively with ribonuclease and proteinase. Treatment with phenol, extraction with chloroform – isoamyl alcohol and precipitation of the plasmid DNA with chilled ethanol followed. In another procedure the yield was much better increased by quickly bringing the lysozyme treated cell suspension to boil, holding at 100 °C for 40 second and then immediately immersing in ice cold water. The chromosomal DNA was removed as usual by centrifugation.
It may be noted here that though the hydrogen bonds between the two strands of duplex DNA are disrupted by alkali or bonds are reformed instantly on restoration of favorable pH or temperature, because of their closed circular nature. Preparations by both these methods, however, contained higher proportion of contaminating linear fragments of chromosomal DNA.
Purification of Plasmid DNA:
Isopycnic Ultracentrifugation:
Plasmid DNA can be purified by equilibrium centrifugation in a Cesium chloride (CsCl) ethidium bromide gradient. Dyes like ethidium bromide and propidium bromide binds to duplex DNA by intercalating between the two strands. This intercalation is much more in case of linear duplex DNA, apparently because of easy access through the two open ends and least with covalent, closed circular super-coiled plasmid DNA.
The bound dye caused a greater reduction in the apparent buoyant density of the linear DNA compared to that of the circular DNA and hence the plasmids could be separated from the linear chromosomal DNA by equilibrium centrifugation the CsCl density gradient, (Fig. 23.2) shows the relative positions of proteins, linear DNA or nicked plasmid DNA, supercoiled plasmid DNA and single stranded RNA.
The plasmid DNA was recovered by puncturing the centrifuge tube with a hypodermic needle (not too fine) just under the plasmid band which could be visualized by low wave length ultraviolet light. The principle behind centrifugal purification has been described.
There are many genetic engineering operations in which very pure plasmid DNA is not required. Various small columns are commercially available which can be effectively used to obtain a considerably pure plasmid DNA preparation from a crude preparation even from a cell lysate. These columns contain among other materials, silica based resin or membrane anion exchange systems, paramagnetic particles or just plain glass powder.
Adsorption chromatography:
In chromatography, Hydroxyapatite is used which binds to the proteins and nucleic acids at low phosphate concentration. If the phosphate concentration is stepwise raised, proteins and RNA are eluted first followed by small RNA molecules i.e. plasmid DNA and finally high molecular weight i.e., chromosomal DNA at the highest phosphate concentration. When clear lysate is loaded onto hydroxyapatite column, separation occurs based on above-mentioned principle.
Preparative electrophoresis:
Plasmid molecule especially in their complete superhelical CCC form can be easily separated from RNA and large chromosomal DNA by electrophoresis in agrose gel. It can yield DNA in mg quantity and can also give an idea of the molecular weight of the DNA by comparing it with the molecular weight marker.
Isolation of RNA:
Cells contain ribo-nucleases and hence inactivating this enzyme during cell lysis is crucial. Agents that effectively inactivate ribo-nucleases are therefore, added during cell lysis. The ionic detergents sodium dodecyl sulphate and sodium lauryl sarkosinate are known to inhibit ribonucleases and are frequently used. Reducing agents like P-merceptoethanol interferes with the tertiary structure of ribonuclease and could be effectively used for inactivating the enzyme.
Once the cell lysate is obtained, RNA is isolated through steps similar to that of DNA isolation. DNA in the cell lysate is shared by repeated ejection through a fine hypodermic needle, proteolytic enzymes added and then the RNA is extracted with phenol or a phenol chloroform mixture.
The next critical step here is to bring the pH down to pH 4.7-5.2 and also to saturate the phenol to be used for extraction with acetate buffer pH 4.7-5.2 instead of Tris buffer pH 8.0. This drives the DNA to the inter-phase of phenol and water and leaves the RNA in the water phase. The RNA is precipitated as usual from the aqueous phase by ethanol or iso-propanol.
Ribosomal RNA constitutes 80-85% of total cellular RNA, transfer RNA and small nuclear RNA (in case of eukaryotes) constitute about 10-15% while messenger RNA constitutes are about 1- 5%. For the most genetic engineering experiments it is the messenger RNA that is important.
Purification of Messenger RNA:
Bacterial messenger RNA can be separated from ribosomal and transfer RNAs by velocity sucrose density gradient centrifugation or gel electrophoresis. Method for purification of eukaryotic mRNA was developed by Philip Leder in 1972 by taking advantage of the fact that mRNA most often has a polytail. The polyA RNA could be separated from other species of RNA by affinity chromatography though a column of oligo (dT) cellulose. A polyU sepharose column has also been used by some.
Analysis of Yield of Nuclei Acid:
Analysis of Yield DNA:
The analysis of yield of purified DNA is best performed by the diphenylamine method. The intensity of the blue color formed at acidic pH is determined by monitoring absorption at 595 nm. This assay is specific and can be used to assay DNA even in a crude DNA preparation.
A quick but less sensitive and much less specific method that is quite often used is to determine absorption of the nucleic acid itself at 260 nm. This assay is based on the fact that the purine and pyrimidine base present in nucleic acids absorb maximally around 260 nm. This assay would obviously be positive for both DNA and RNA, free nucleotides and nucleosides and even purine and pyrimidine based, but in a purified DNA or RNA sample the assay is reliable. Another quick but very approximate assay of double stranded DNA is by visual estimation of the ultraviolet fluorescence of DNA in a droplet elicited by ethidium bromide.
Analysis of Yield RNA:
It is obvious from the preceding subsection that yield of RNA can also be determined by monitoring absorption at 260 nm as long as it is pure sample of RNA. A more specific and sensitive method is based on the monitoring of the color developed by reaction with orcinol. The RNA sample is boiled with freshly re-crystallized orcinol in the presence of FeCl and HCl.
The green color, developed is monitored at 655 nm. Ribose linked only to purines contributes to the color. Specificity of this reaction is not as good as the diphenyl amine reaction for DNA 2- Deoxyribose DNA methyl pentose, hexuronic acid and aldoheptoses also produce some green colour giving absorbance in that region.
Method # 3. DNA Sequencing:
DNA sequencing is the determination of the precise sequence of nucleotides in a sample of DNA. Before the development of direct DNA sequencing methods, DNA sequencing was difficult and indirect. The DNA had to be converted to RNA, and limited RNA sequencing could be done by the existing cumbersome methods. Thus, only shorter DNA sequences could be determined by this method. Using this method, Walter Gilbert and Alan Maxam at Havard University determined that the Lac operator is a 27 bp long sequence.
The development of direct DNA sequencing techniques changed the scope of biological research. The evolution of DNA sequencing technology from plus-minus sequencing to pyro-sequencing within about 20 years parallels the progress in biology from molecular biology to genomics.
The development of DNA sequencing techniques with enhanced speed, sensitivity and throughput are of utmost importance for the study of biological systems. Sequence determination is most commonly performed using di-deoxy chain termination technology. Pyro-sequencing, a non-electrophoretic real- time bio-luminometric method for DNA sequencing has emerged as a state of the art sequencing technology.
This technology has the advantage of accuracy, ease of use, and high flexibility for different applications. Pyro-sequencing allows the analysis of genetic variations including SNPs, insertion/deletions and short repeats, as well as assessing RNA allelic imbalance, DNA methylation status and gene copy number.