In this article we will discuss about:- 1. Introduction to Gene Interaction 2. Allelic Gene Interactions 3. Non Allelic 4. Multiple Factors and Polygenic 5. Other Kinds.
Contents:
- Introduction to Gene Interaction
- Allelic Gene Interactions
- Non Allelic Gene Interactions
- Multiple Factors and Polygenic Gene Interactions
- Other Kinds of Gene Interactions
1. Introduction to Gene Interaction:
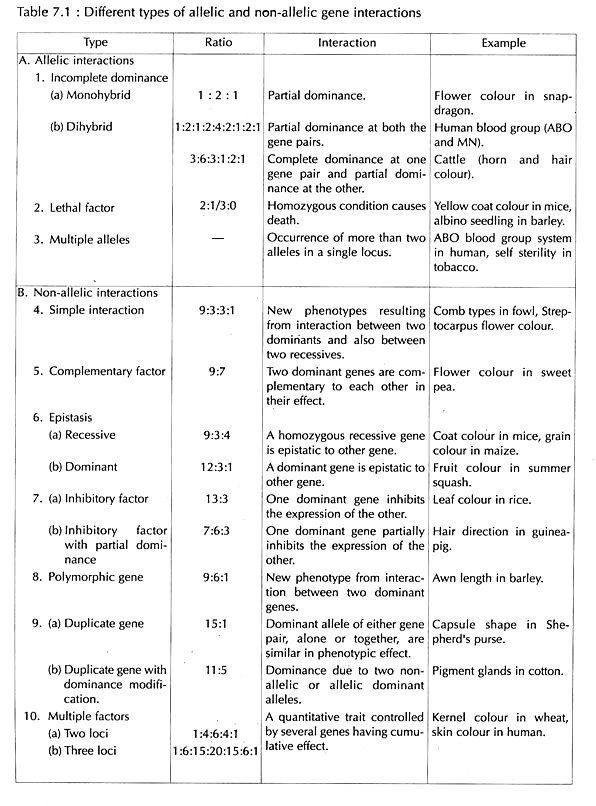
Mendelian genetics does not explain all kinds of inheritance for which the phenotypic ratios in some cases are different from Mendelian ratios (3:1 for monohybrid, 9:3:3:1 for di-hybrid in F2). This is because sometimes a particular allele may be partially or equally dominant to the other or due to existence of more than two alleles or due to lethal alleles. These kinds of genetic interactions between the alleles of a single gene are referred to as allelic or intra- allelic interactions.
Non-allelic or inter-allelic interactions also occur where the development of single character is due to two or more genes affecting the expression of each other in various ways.
Thus, the expression of gene is not independent of each other and dependent on the presence or absence of other gene or genes; These kinds of deviations from Mendelian one gene-one trait concept is known as Factor Hypothesis or Interaction of Genes (Table 7.1).
2. Allelic Gene Interactions:
Incomplete Dominance or Blending Inheritance (1:2:1):
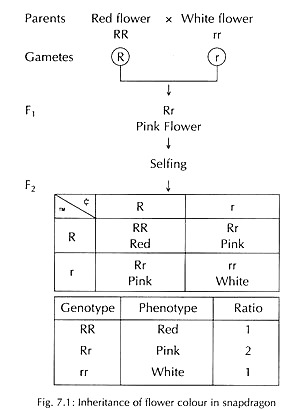
A dominant allele may not completely suppress other allele, hence a heterozygote is phenotypically distinguishable (intermediate phenotype) from either homozygotes.
In snapdragon and Mirabilis jalapa, the cross between pure bred red-flowered and white-flowered plants yields pink-flowered F1 hybrid plants (deviation from parental phenotypes), i.e., intermediate of the two parents. When F1 plants are self-fertilized, the F2 progeny shows three classes of plants in the ratio 1 red: 2 pink: 1 white instead of 3:1 (Fig. 7.1).
Therefore, a F1 di-hybrid showing incomplete dominance for both the characters will segregate in F2 into (1 :2 : 1) X (1 :2 : 1 ) = 1 :2 : 1 : 2 : 4 : 2:1 : 2 : 1. And a F1 di-hybrid showing complete dominance for one trait and incomplete dominance of another trait will segregate in F2 into (3:1) x (1 :2:1) = 3:6:3:1 :2:1.
Co-dominance:
Here both the alleles of a gene express themselves in the heterozygotes. Phenotypes of both the parents appear in F1 hybrid rather than the intermediate phenotype. In human, MN blood group is controlled by a single gene.
Only two alleles exist, M and N. Father with N blood group (genotype NN) and mother with M blood group (genotype MM) will have children with MN blood group (genotype MN). Both phenotypes are identifiable in the hybrid. F2 segregates in the ratio 1M blood group: 2 MN blood group : 1 N blood group.
Over-dominance:
Sometimes the phenotype of F1 heterozygote is more extreme than that of either parents. The amount of fluorescent eye pigment in heterozygous white eyes of Drosophila exceeds that found in either parents.
Lethal Factor (2:1):
The genes which cause the death of the individual carrying it, is called lethal factor. Recessive lethals are expressed only when they are in homozygous state and the heterozygotes remain unaffected. There are genes that have a dominant phenotypic effect but are recessive lethal, e.g., gene for yellow coat colour in mice.
But many genes are recessive both in their phenotypic as well as lethal effects, e.g., gene producing albino seedlings in barley (Fig. 7.2).
Dominant lethals are lost from the population because they cause death of the organism even in a heterozygous state, e.g., epiloia gene in human beings. Conditional lethals require a specific condition for their lethal action, e.g., temperature sensitive mutant of barley (lethal effect at low temperature).
Balanced lethals are all heterozygous for the lethal genes; both dominant and recessive homozygotes will die, e.g., balanced lethal system in Oenothera. Gametic lethals make the gametes incapable of fertilization, e.g., segregation distorter gene in male Drosophila.
Semi-lethal genes do not cause the death of all the individuals, e.g., xentha mutants in some plants.
Multiple Alleles:
A gene for particular character may have more than two allelomorphs or alleles occupying same locus of the chromosome (only two of them present in a diploid organism). These allelomorphs make a series of multiple alleles.
Human ABO blood group system furnishes best example. The gene for antigen may occur in three possible allelic forms – lA, IB, i. The allele for the A antigen is co-dominant with the allele I8 for the B antigen. Both are completely dominant to the allele i which fails to specify any detectable antigenic structure. Therefore, the possible genotypes of the four blood groups are shown in Fig. 7.3.
Self-sterility in tobacco is determined by the gene with many different allelic forms. If there are only three alleles (s1, s2, s3), the possible genotypes of plants are s1s2, s1s3, s2s3 (always heterozygous), homozygous genotypes (s1s1, s2s2, s3s3) are not possible in a self-incompatible species.
In this case, pollen carrying an allele different from the two alleles present in the female plant will be able to function resulting in restriction of fertility (Fig 7.4).Iso-alleles express themselves within the same phenotypic range, e.g., in Drosophila several alleles (W+s ,W+c, W+g) exhibit red eye colour.
3. Non Allelic Gene Interactions:
Simple Interaction (9:3:3:1):
In this case, two non-alleiic gene pairs affect the same character. The dominant allele of each of the two factors produces separate phenotypes when they are alone. When both the dominant alleles are present together, they produce a distinct new phenotype. The absence of both the dominant alleles gives rise to yet another phenotype.
The inheritance of comb types in fowls is the best example where R gene gives rise to rose comb and P gene gives rise to pea comb; both are dominant over single comb; the presence of both the dominant genes results in walnut comb (Fig. 7.5). Similar pattern of inheritance is found in Streptocarpus flower colour (Fig. 7.6).
Complementary Factor (9:7):
Certain characters are produced by the interaction between two or more genes occupying different loci inherited from different parents. These genes are complementary to one another, i.e., if present alone they remain unexpressed, only when they are brought together through suitable crossing will express.
In sweet pea (Lathyrus odoratus), both the genes C and P are required to synthesize anthocyanin pigment causing purple colour. But absence of any one cannot produce anthocyanin causing white flower. So C and P are complementary to each other for anthocyanin formation (Fig. 7.7).
Involvement of more than two complementary genes is possible, e.g., three complementary genes governing aleurone colour in maize.
Epistasis:
When a gene or gene pair masks or prevents the expression of other non-allelic gene, called epistasis. The gene which produces the effect called epistatic gene and the gene whose expression is suppressed called hypostatic gene.
(a) Recessive Epistasis or Supplementary Factor (9:3:4):
In this case, homozygous recessive condition of a gene determines the phenotype irrespective of the alleles of other gene pairs, i.e., recessive allele hides the effect of the other gene. The coat colour of mice is controlled by two pairs of genes.
Dominant gene C produces black colour, absence of it causes albino. Gene A produces agouti colour in presence of C, but cannot express in absence of it (with cc) resulting in albino. Thus recessive allele c (cc) is epistatic to dominant allele A (Fig. 7.8).
The grain colour in maize is governed by two genes — R (red) and Pr (purple). The recessive allele rr is epistatic to gene Pr (Fig. 7.9).
(b) Dominant Epistasis (12:3:1):
Sometimes a dominant gene does not allow the expression of other non-allelic gene called dominant epistasis. In summer squash, the fruit colour is governed by two genes. The dominant gene W for white colour, suppresses the expression of the gene Y which controls yellow colour. So yellow colour appears only in absence of W. Thus W is epistatic to Y. In absence of both W and Y, green colour develops (Fig. 7.10).
Inhibitory Factor:
Inhibitory factor is such a gene which itself has no phenotypic effect but inhibits the expression of another non-allelic gene; in rice, purple leaf colour is due to gene P, and p causing green colour. Another non-allelic dominant gene I inhibits the expression of P but is ineffective in recessive form (ii). Thus the factor I has no visible effect of its own but inhibits the colour expression of P (Fig. 7.11).
Inhibitory Factor with Partial Dominance (7:6:3):
Sometimes an inhibitory gene shows incomplete dominance thus allowing the expression of other gene partially. In guineapig, hair direction is controlled by two genes. Rough (R) hair is dominant over smooth (r) hair, other gene I is inhibitory to R at horinozygous state (II) but in heterozygous state (II) causes partially rough (Fig. 7.12).
Polymorphic Gene (9:6:1):
Here two non-allelilc genes controlling a character produce identical phenotype when they are alone, but when both the genes are present together their phenotypes effect is enhanced due to cumulative effect. In barley, two genes A and B affect the length of awns.
Gene A or B alone gives rise to awns of medium length (the effect of A is same as B); but when both present, long awn is produced; absence of both results aweless (Fig. 7.13).
Duplicate Gene (15:1):
Sometimes a character is controlled by two non-allelic genes whose dominant alleles produce the same phenotype whether they are alone or together. In Shepherd’s purse (Capsella bursa-pastoris), the presence of either gene A or gene B or both results in triangular capsules; when both these genes are in recessive forms, the oval capsules produced (Fig. 7.14).
Duplicate Gene with Dominance Modification (11:5):
A character controlled by two gene pairs showing dominance only if two dominant alleles are present. Dominant phenotype will thus be produced only when two non-allelic dominant alleles or two allelic dominant alleles are present. Such a case is found in pigment glands of cotton (Fig. 7.15).
4. Multiple Factors and Polygenic Gene Inheritance:
Though some characters (qualitative) show discontinuous variation but a majority of characters (quantitative, e.g., height, weight, etc.) exhibit continuous variation. Yule, Nilsson-Ehle, East suggested that quantitative variation is controlled by large number of individual genes called polygenic systems and the inheritance could be explained on the basis of multiple factor hypothesis.
The hypothesis states that for a given quantitative trait there could be several genes, which are independent in their segregation and had cumulative effect on the phenotype.
Kernel colour in wheat is a quantitative character and controlled by two different genes. The heterozygote is intermediate in colour between the two homozygotes. Both the dominant genes have small and equal (or almost equal) effects on seed colour. F1 heterozygote for two genes will segregate in F2 in the ratio 1:4: 6:4:1.
The intensity of seed colour depends on the number of dominant alleles present, i.e., their effects are cumulative in nature (Fig. 7.16). It is now known that there are three genes involved in kernel colour in wheat, thus a F, heterozygous for all three genes will segregate in F1 in the ratio 1 : 6 : 15 : 20 : 15 : 6 : 1.
Corolla length in Nicotiana is another quantitative character. The crosses made between two inbred varieties of N. longiflora or N. tabaccum (tobacco) differing in corolla length, show F1 with uniform corolla length but F2 exhibits a greater degree of variation.
Mean value of F3 derived from single F2 plant with particular corolla length differs greatly from other single plant F3 progenies. It is thus obvious that F2 plants differ genetically (Fig. 7.17).
Skin colour in human beings is under polygenic effect, the number of gene pairs involved may be two (Fig. 7.18) or more than two, possibly four or five.
The number of genes involved in polygenic inheritance can be calculated from the frequency of parental type using the formula 1/4n (n = number of gene pairs).
If parental type obtained is one out of every 64 offspring’s (1/64), then the number of genes involved will be three (4n = 64 = 43).
5. Other Kinds of Gene Interactions:
Modifiers:
Genes which modify the phenotypic effect of a major gene called modifying gene. They reduce or enhance the effect of other gene in quantitative manner, e.g., genes responsible for dilution of body colour.
Suppressors:
Genes which will not allow mutant allele of another gene to express resulting in wild phenotype called suppressor gene, e.g., Su-s in Drosophila suppresses the expression of dominant mutant gene star eye(s).
Pleiotropy:
Gene having more than one effect (multiple effects) are called pleiotropic genes. They have a major effect in addition to secondary effect. In Drosophila, the genes for bristle, eye and wing significantly influence the number of facets in bar-eyed individuals.
Atavism:
The appearance of offspring’s which resemble their remote ancestors called atavism.
Penetrance:
The ability of a gene to be expressed phenotypically to any degree is called penetrance. Penetrance may be complete, e.g., in pea, expression of R allele for red flower in homozygous and heterozygous conditions. It may be incomplete, e.g., dominant gene P for Polydactyly in human, sometimes does not show polydactylous condition in heterozygous state.
Expressivity:
A trait though penetrant, may be quite variable in its phenotypic expression, e.g., in human polydactylous condition may be penetrant in left hand but not in the right.